Merge branch 'develop' into check-isam
commit
650bd576ea
|
|
@ -122,21 +122,13 @@ using MotionModel = BetweenFactor<double>;
|
||||||
// Test fixture with switching network.
|
// Test fixture with switching network.
|
||||||
/// ϕ(X(0)) .. ϕ(X(k),X(k+1)) .. ϕ(X(k);z_k) .. ϕ(M(0)) .. ϕ(M(K-3),M(K-2))
|
/// ϕ(X(0)) .. ϕ(X(k),X(k+1)) .. ϕ(X(k);z_k) .. ϕ(M(0)) .. ϕ(M(K-3),M(K-2))
|
||||||
struct Switching {
|
struct Switching {
|
||||||
private:
|
|
||||||
HybridNonlinearFactorGraph nonlinearFactorGraph_;
|
|
||||||
|
|
||||||
public:
|
public:
|
||||||
size_t K;
|
size_t K;
|
||||||
DiscreteKeys modes;
|
DiscreteKeys modes;
|
||||||
HybridNonlinearFactorGraph unaryFactors, binaryFactors, modeChain;
|
HybridNonlinearFactorGraph unaryFactors, binaryFactors, modeChain;
|
||||||
HybridGaussianFactorGraph linearizedFactorGraph;
|
HybridGaussianFactorGraph linearUnaryFactors, linearBinaryFactors;
|
||||||
Values linearizationPoint;
|
Values linearizationPoint;
|
||||||
|
|
||||||
// Access the flat nonlinear factor graph.
|
|
||||||
const HybridNonlinearFactorGraph &nonlinearFactorGraph() const {
|
|
||||||
return nonlinearFactorGraph_;
|
|
||||||
}
|
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* @brief Create with given number of time steps.
|
* @brief Create with given number of time steps.
|
||||||
*
|
*
|
||||||
|
|
@ -166,37 +158,33 @@ struct Switching {
|
||||||
// Create hybrid factor graph.
|
// Create hybrid factor graph.
|
||||||
|
|
||||||
// Add a prior ϕ(X(0)) on X(0).
|
// Add a prior ϕ(X(0)) on X(0).
|
||||||
nonlinearFactorGraph_.emplace_shared<PriorFactor<double>>(
|
unaryFactors.emplace_shared<PriorFactor<double>>(
|
||||||
X(0), measurements.at(0), Isotropic::Sigma(1, prior_sigma));
|
X(0), measurements.at(0), Isotropic::Sigma(1, prior_sigma));
|
||||||
unaryFactors.push_back(nonlinearFactorGraph_.back());
|
|
||||||
|
|
||||||
// Add "motion models" ϕ(X(k),X(k+1),M(k)).
|
// Add "motion models" ϕ(X(k),X(k+1),M(k)).
|
||||||
for (size_t k = 0; k < K - 1; k++) {
|
for (size_t k = 0; k < K - 1; k++) {
|
||||||
auto motion_models = motionModels(k, between_sigma);
|
auto motion_models = motionModels(k, between_sigma);
|
||||||
nonlinearFactorGraph_.emplace_shared<HybridNonlinearFactor>(
|
binaryFactors.emplace_shared<HybridNonlinearFactor>(modes[k],
|
||||||
modes[k], motion_models);
|
motion_models);
|
||||||
binaryFactors.push_back(nonlinearFactorGraph_.back());
|
|
||||||
}
|
}
|
||||||
|
|
||||||
// Add measurement factors ϕ(X(k);z_k).
|
// Add measurement factors ϕ(X(k);z_k).
|
||||||
auto measurement_noise = Isotropic::Sigma(1, prior_sigma);
|
auto measurement_noise = Isotropic::Sigma(1, prior_sigma);
|
||||||
for (size_t k = 1; k < K; k++) {
|
for (size_t k = 1; k < K; k++) {
|
||||||
nonlinearFactorGraph_.emplace_shared<PriorFactor<double>>(
|
unaryFactors.emplace_shared<PriorFactor<double>>(X(k), measurements.at(k),
|
||||||
X(k), measurements.at(k), measurement_noise);
|
measurement_noise);
|
||||||
unaryFactors.push_back(nonlinearFactorGraph_.back());
|
|
||||||
}
|
}
|
||||||
|
|
||||||
// Add "mode chain" ϕ(M(0)) ϕ(M(0),M(1)) ... ϕ(M(K-3),M(K-2))
|
// Add "mode chain" ϕ(M(0)) ϕ(M(0),M(1)) ... ϕ(M(K-3),M(K-2))
|
||||||
modeChain = createModeChain(transitionProbabilityTable);
|
modeChain = createModeChain(transitionProbabilityTable);
|
||||||
nonlinearFactorGraph_ += modeChain;
|
|
||||||
|
|
||||||
// Create the linearization point.
|
// Create the linearization point.
|
||||||
for (size_t k = 0; k < K; k++) {
|
for (size_t k = 0; k < K; k++) {
|
||||||
linearizationPoint.insert<double>(X(k), static_cast<double>(k + 1));
|
linearizationPoint.insert<double>(X(k), static_cast<double>(k + 1));
|
||||||
}
|
}
|
||||||
|
|
||||||
linearizedFactorGraph =
|
linearUnaryFactors = *unaryFactors.linearize(linearizationPoint);
|
||||||
*nonlinearFactorGraph_.linearize(linearizationPoint);
|
linearBinaryFactors = *binaryFactors.linearize(linearizationPoint);
|
||||||
}
|
}
|
||||||
|
|
||||||
// Create motion models for a given time step
|
// Create motion models for a given time step
|
||||||
|
|
@ -227,6 +215,24 @@ struct Switching {
|
||||||
}
|
}
|
||||||
return chain;
|
return chain;
|
||||||
}
|
}
|
||||||
|
|
||||||
|
/// Get the full linear factor graph.
|
||||||
|
HybridGaussianFactorGraph linearizedFactorGraph() const {
|
||||||
|
HybridGaussianFactorGraph graph;
|
||||||
|
graph.push_back(linearUnaryFactors);
|
||||||
|
graph.push_back(linearBinaryFactors);
|
||||||
|
graph.push_back(modeChain);
|
||||||
|
return graph;
|
||||||
|
}
|
||||||
|
|
||||||
|
/// Get all the nonlinear factors.
|
||||||
|
HybridNonlinearFactorGraph nonlinearFactorGraph() const {
|
||||||
|
HybridNonlinearFactorGraph graph;
|
||||||
|
graph.push_back(unaryFactors);
|
||||||
|
graph.push_back(binaryFactors);
|
||||||
|
graph.push_back(modeChain);
|
||||||
|
return graph;
|
||||||
|
}
|
||||||
};
|
};
|
||||||
|
|
||||||
} // namespace gtsam
|
} // namespace gtsam
|
||||||
|
|
|
||||||
|
|
@ -31,6 +31,7 @@
|
||||||
|
|
||||||
// Include for test suite
|
// Include for test suite
|
||||||
#include <CppUnitLite/TestHarness.h>
|
#include <CppUnitLite/TestHarness.h>
|
||||||
|
|
||||||
#include <memory>
|
#include <memory>
|
||||||
|
|
||||||
using namespace std;
|
using namespace std;
|
||||||
|
|
@ -263,7 +264,7 @@ TEST(HybridBayesNet, Choose) {
|
||||||
const Ordering ordering(s.linearizationPoint.keys());
|
const Ordering ordering(s.linearizationPoint.keys());
|
||||||
|
|
||||||
const auto [hybridBayesNet, remainingFactorGraph] =
|
const auto [hybridBayesNet, remainingFactorGraph] =
|
||||||
s.linearizedFactorGraph.eliminatePartialSequential(ordering);
|
s.linearizedFactorGraph().eliminatePartialSequential(ordering);
|
||||||
|
|
||||||
DiscreteValues assignment;
|
DiscreteValues assignment;
|
||||||
assignment[M(0)] = 1;
|
assignment[M(0)] = 1;
|
||||||
|
|
@ -292,7 +293,7 @@ TEST(HybridBayesNet, OptimizeAssignment) {
|
||||||
const Ordering ordering(s.linearizationPoint.keys());
|
const Ordering ordering(s.linearizationPoint.keys());
|
||||||
|
|
||||||
const auto [hybridBayesNet, remainingFactorGraph] =
|
const auto [hybridBayesNet, remainingFactorGraph] =
|
||||||
s.linearizedFactorGraph.eliminatePartialSequential(ordering);
|
s.linearizedFactorGraph().eliminatePartialSequential(ordering);
|
||||||
|
|
||||||
DiscreteValues assignment;
|
DiscreteValues assignment;
|
||||||
assignment[M(0)] = 1;
|
assignment[M(0)] = 1;
|
||||||
|
|
@ -319,7 +320,7 @@ TEST(HybridBayesNet, Optimize) {
|
||||||
Switching s(4, 1.0, 0.1, {0, 1, 2, 3}, "1/1 1/1");
|
Switching s(4, 1.0, 0.1, {0, 1, 2, 3}, "1/1 1/1");
|
||||||
|
|
||||||
HybridBayesNet::shared_ptr hybridBayesNet =
|
HybridBayesNet::shared_ptr hybridBayesNet =
|
||||||
s.linearizedFactorGraph.eliminateSequential();
|
s.linearizedFactorGraph().eliminateSequential();
|
||||||
|
|
||||||
HybridValues delta = hybridBayesNet->optimize();
|
HybridValues delta = hybridBayesNet->optimize();
|
||||||
|
|
||||||
|
|
@ -347,7 +348,7 @@ TEST(HybridBayesNet, Pruning) {
|
||||||
Switching s(3);
|
Switching s(3);
|
||||||
|
|
||||||
HybridBayesNet::shared_ptr posterior =
|
HybridBayesNet::shared_ptr posterior =
|
||||||
s.linearizedFactorGraph.eliminateSequential();
|
s.linearizedFactorGraph().eliminateSequential();
|
||||||
EXPECT_LONGS_EQUAL(5, posterior->size());
|
EXPECT_LONGS_EQUAL(5, posterior->size());
|
||||||
|
|
||||||
// Optimize
|
// Optimize
|
||||||
|
|
@ -400,7 +401,7 @@ TEST(HybridBayesNet, Prune) {
|
||||||
Switching s(4);
|
Switching s(4);
|
||||||
|
|
||||||
HybridBayesNet::shared_ptr posterior =
|
HybridBayesNet::shared_ptr posterior =
|
||||||
s.linearizedFactorGraph.eliminateSequential();
|
s.linearizedFactorGraph().eliminateSequential();
|
||||||
EXPECT_LONGS_EQUAL(7, posterior->size());
|
EXPECT_LONGS_EQUAL(7, posterior->size());
|
||||||
|
|
||||||
HybridValues delta = posterior->optimize();
|
HybridValues delta = posterior->optimize();
|
||||||
|
|
@ -418,7 +419,7 @@ TEST(HybridBayesNet, UpdateDiscreteConditionals) {
|
||||||
Switching s(4);
|
Switching s(4);
|
||||||
|
|
||||||
HybridBayesNet::shared_ptr posterior =
|
HybridBayesNet::shared_ptr posterior =
|
||||||
s.linearizedFactorGraph.eliminateSequential();
|
s.linearizedFactorGraph().eliminateSequential();
|
||||||
EXPECT_LONGS_EQUAL(7, posterior->size());
|
EXPECT_LONGS_EQUAL(7, posterior->size());
|
||||||
|
|
||||||
DiscreteConditional joint;
|
DiscreteConditional joint;
|
||||||
|
|
|
||||||
|
|
@ -337,7 +337,7 @@ TEST(HybridBayesTree, OptimizeMultifrontal) {
|
||||||
Switching s(4);
|
Switching s(4);
|
||||||
|
|
||||||
HybridBayesTree::shared_ptr hybridBayesTree =
|
HybridBayesTree::shared_ptr hybridBayesTree =
|
||||||
s.linearizedFactorGraph.eliminateMultifrontal();
|
s.linearizedFactorGraph().eliminateMultifrontal();
|
||||||
HybridValues delta = hybridBayesTree->optimize();
|
HybridValues delta = hybridBayesTree->optimize();
|
||||||
|
|
||||||
VectorValues expectedValues;
|
VectorValues expectedValues;
|
||||||
|
|
@ -349,30 +349,40 @@ TEST(HybridBayesTree, OptimizeMultifrontal) {
|
||||||
EXPECT(assert_equal(expectedValues, delta.continuous(), 1e-5));
|
EXPECT(assert_equal(expectedValues, delta.continuous(), 1e-5));
|
||||||
}
|
}
|
||||||
|
|
||||||
|
namespace optimize_fixture {
|
||||||
|
HybridGaussianFactorGraph GetGaussianFactorGraph(size_t N) {
|
||||||
|
Switching s(N);
|
||||||
|
HybridGaussianFactorGraph graph;
|
||||||
|
|
||||||
|
for (size_t i = 0; i < N - 1; i++) {
|
||||||
|
graph.push_back(s.linearBinaryFactors.at(i));
|
||||||
|
}
|
||||||
|
for (size_t i = 0; i < N; i++) {
|
||||||
|
graph.push_back(s.linearUnaryFactors.at(i));
|
||||||
|
}
|
||||||
|
for (size_t i = 0; i < N - 1; i++) {
|
||||||
|
graph.push_back(s.modeChain.at(i));
|
||||||
|
}
|
||||||
|
|
||||||
|
return graph;
|
||||||
|
}
|
||||||
|
} // namespace optimize_fixture
|
||||||
|
|
||||||
/* ****************************************************************************/
|
/* ****************************************************************************/
|
||||||
// Test for optimizing a HybridBayesTree with a given assignment.
|
// Test for optimizing a HybridBayesTree with a given assignment.
|
||||||
TEST(HybridBayesTree, OptimizeAssignment) {
|
TEST(HybridBayesTree, OptimizeAssignment) {
|
||||||
Switching s(4);
|
using namespace optimize_fixture;
|
||||||
|
|
||||||
|
size_t N = 4;
|
||||||
|
|
||||||
HybridGaussianISAM isam;
|
HybridGaussianISAM isam;
|
||||||
HybridGaussianFactorGraph graph1;
|
|
||||||
|
|
||||||
// Add the 3 hybrid factors, x1-x2, x2-x3, x3-x4
|
// Add the 3 hybrid factors, x0-x1, x1-x2, x2-x3
|
||||||
for (size_t i = 1; i < 4; i++) {
|
// Then add the Gaussian factors, 1 prior on X(0),
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(i));
|
// 3 measurements on X(1), X(2), X(3)
|
||||||
}
|
// Finally add the discrete factors
|
||||||
|
// m0, m1-m0, m2-m1
|
||||||
// Add the Gaussian factors, 1 prior on X(1),
|
HybridGaussianFactorGraph graph1 = GetGaussianFactorGraph(N);
|
||||||
// 3 measurements on X(2), X(3), X(4)
|
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(0));
|
|
||||||
for (size_t i = 4; i <= 7; i++) {
|
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(i));
|
|
||||||
}
|
|
||||||
|
|
||||||
// Add the discrete factors
|
|
||||||
for (size_t i = 7; i <= 9; i++) {
|
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(i));
|
|
||||||
}
|
|
||||||
|
|
||||||
isam.update(graph1);
|
isam.update(graph1);
|
||||||
|
|
||||||
|
|
@ -394,12 +404,13 @@ TEST(HybridBayesTree, OptimizeAssignment) {
|
||||||
|
|
||||||
EXPECT(assert_equal(expected_delta, delta));
|
EXPECT(assert_equal(expected_delta, delta));
|
||||||
|
|
||||||
|
Switching s(N);
|
||||||
// Create ordering.
|
// Create ordering.
|
||||||
Ordering ordering;
|
Ordering ordering;
|
||||||
for (size_t k = 0; k < s.K; k++) ordering.push_back(X(k));
|
for (size_t k = 0; k < s.K; k++) ordering.push_back(X(k));
|
||||||
|
|
||||||
const auto [hybridBayesNet, remainingFactorGraph] =
|
const auto [hybridBayesNet, remainingFactorGraph] =
|
||||||
s.linearizedFactorGraph.eliminatePartialSequential(ordering);
|
s.linearizedFactorGraph().eliminatePartialSequential(ordering);
|
||||||
|
|
||||||
GaussianBayesNet gbn = hybridBayesNet->choose(assignment);
|
GaussianBayesNet gbn = hybridBayesNet->choose(assignment);
|
||||||
VectorValues expected = gbn.optimize();
|
VectorValues expected = gbn.optimize();
|
||||||
|
|
@ -410,38 +421,29 @@ TEST(HybridBayesTree, OptimizeAssignment) {
|
||||||
/* ****************************************************************************/
|
/* ****************************************************************************/
|
||||||
// Test for optimizing a HybridBayesTree.
|
// Test for optimizing a HybridBayesTree.
|
||||||
TEST(HybridBayesTree, Optimize) {
|
TEST(HybridBayesTree, Optimize) {
|
||||||
Switching s(4);
|
using namespace optimize_fixture;
|
||||||
|
|
||||||
|
size_t N = 4;
|
||||||
|
|
||||||
HybridGaussianISAM isam;
|
HybridGaussianISAM isam;
|
||||||
HybridGaussianFactorGraph graph1;
|
// Add the 3 hybrid factors, x0-x1, x1-x2, x2-x3
|
||||||
|
// Then add the Gaussian factors, 1 prior on X(0),
|
||||||
// Add the 3 hybrid factors, x1-x2, x2-x3, x3-x4
|
// 3 measurements on X(1), X(2), X(3)
|
||||||
for (size_t i = 1; i < 4; i++) {
|
// Finally add the discrete factors
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(i));
|
// m0, m1-m0, m2-m1
|
||||||
}
|
HybridGaussianFactorGraph graph1 = GetGaussianFactorGraph(N);
|
||||||
|
|
||||||
// Add the Gaussian factors, 1 prior on X(0),
|
|
||||||
// 3 measurements on X(2), X(3), X(4)
|
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(0));
|
|
||||||
for (size_t i = 4; i <= 6; i++) {
|
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(i));
|
|
||||||
}
|
|
||||||
|
|
||||||
// Add the discrete factors
|
|
||||||
for (size_t i = 7; i <= 9; i++) {
|
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(i));
|
|
||||||
}
|
|
||||||
|
|
||||||
isam.update(graph1);
|
isam.update(graph1);
|
||||||
|
|
||||||
HybridValues delta = isam.optimize();
|
HybridValues delta = isam.optimize();
|
||||||
|
|
||||||
|
Switching s(N);
|
||||||
// Create ordering.
|
// Create ordering.
|
||||||
Ordering ordering;
|
Ordering ordering;
|
||||||
for (size_t k = 0; k < s.K; k++) ordering.push_back(X(k));
|
for (size_t k = 0; k < s.K; k++) ordering.push_back(X(k));
|
||||||
|
|
||||||
const auto [hybridBayesNet, remainingFactorGraph] =
|
const auto [hybridBayesNet, remainingFactorGraph] =
|
||||||
s.linearizedFactorGraph.eliminatePartialSequential(ordering);
|
s.linearizedFactorGraph().eliminatePartialSequential(ordering);
|
||||||
|
|
||||||
DiscreteFactorGraph dfg = remainingFactorGraph->discreteFactors();
|
DiscreteFactorGraph dfg = remainingFactorGraph->discreteFactors();
|
||||||
|
|
||||||
|
|
@ -461,27 +463,18 @@ TEST(HybridBayesTree, Optimize) {
|
||||||
/* ****************************************************************************/
|
/* ****************************************************************************/
|
||||||
// Test for choosing a GaussianBayesTree from a HybridBayesTree.
|
// Test for choosing a GaussianBayesTree from a HybridBayesTree.
|
||||||
TEST(HybridBayesTree, Choose) {
|
TEST(HybridBayesTree, Choose) {
|
||||||
Switching s(4);
|
using namespace optimize_fixture;
|
||||||
|
|
||||||
|
size_t N = 4;
|
||||||
|
|
||||||
HybridGaussianISAM isam;
|
HybridGaussianISAM isam;
|
||||||
HybridGaussianFactorGraph graph1;
|
|
||||||
|
|
||||||
// Add the 3 hybrid factors, x1-x2, x2-x3, x3-x4
|
// Add the 3 hybrid factors, x0-x1, x1-x2, x2-x3
|
||||||
for (size_t i = 1; i < 4; i++) {
|
// Then add the Gaussian factors, 1 prior on X(0),
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(i));
|
// 3 measurements on X(1), X(2), X(3)
|
||||||
}
|
// Finally add the discrete factors
|
||||||
|
// m0, m1-m0, m2-m1
|
||||||
// Add the Gaussian factors, 1 prior on X(0),
|
HybridGaussianFactorGraph graph1 = GetGaussianFactorGraph(N);
|
||||||
// 3 measurements on X(2), X(3), X(4)
|
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(0));
|
|
||||||
for (size_t i = 4; i <= 6; i++) {
|
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(i));
|
|
||||||
}
|
|
||||||
|
|
||||||
// Add the discrete factors
|
|
||||||
for (size_t i = 7; i <= 9; i++) {
|
|
||||||
graph1.push_back(s.linearizedFactorGraph.at(i));
|
|
||||||
}
|
|
||||||
|
|
||||||
isam.update(graph1);
|
isam.update(graph1);
|
||||||
|
|
||||||
|
|
@ -493,8 +486,9 @@ TEST(HybridBayesTree, Choose) {
|
||||||
GaussianBayesTree gbt = isam.choose(assignment);
|
GaussianBayesTree gbt = isam.choose(assignment);
|
||||||
|
|
||||||
// Specify ordering so it matches that of HybridGaussianISAM.
|
// Specify ordering so it matches that of HybridGaussianISAM.
|
||||||
|
Switching s(N);
|
||||||
Ordering ordering(KeyVector{X(0), X(1), X(2), X(3), M(0), M(1), M(2)});
|
Ordering ordering(KeyVector{X(0), X(1), X(2), X(3), M(0), M(1), M(2)});
|
||||||
auto bayesTree = s.linearizedFactorGraph.eliminateMultifrontal(ordering);
|
auto bayesTree = s.linearizedFactorGraph().eliminateMultifrontal(ordering);
|
||||||
|
|
||||||
auto expected_gbt = bayesTree->choose(assignment);
|
auto expected_gbt = bayesTree->choose(assignment);
|
||||||
|
|
||||||
|
|
|
||||||
|
|
@ -82,7 +82,7 @@ TEST(HybridEstimation, Full) {
|
||||||
// Switching example of robot moving in 1D
|
// Switching example of robot moving in 1D
|
||||||
// with given measurements and equal mode priors.
|
// with given measurements and equal mode priors.
|
||||||
Switching switching(K, 1.0, 0.1, measurements, "1/1 1/1");
|
Switching switching(K, 1.0, 0.1, measurements, "1/1 1/1");
|
||||||
HybridGaussianFactorGraph graph = switching.linearizedFactorGraph;
|
HybridGaussianFactorGraph graph = switching.linearizedFactorGraph();
|
||||||
|
|
||||||
Ordering hybridOrdering;
|
Ordering hybridOrdering;
|
||||||
for (size_t k = 0; k < K; k++) {
|
for (size_t k = 0; k < K; k++) {
|
||||||
|
|
@ -325,7 +325,7 @@ TEST(HybridEstimation, Probability) {
|
||||||
// given measurements and equal mode priors.
|
// given measurements and equal mode priors.
|
||||||
Switching switching(K, between_sigma, measurement_sigma, measurements,
|
Switching switching(K, between_sigma, measurement_sigma, measurements,
|
||||||
"1/1 1/1");
|
"1/1 1/1");
|
||||||
auto graph = switching.linearizedFactorGraph;
|
auto graph = switching.linearizedFactorGraph();
|
||||||
|
|
||||||
// Continuous elimination
|
// Continuous elimination
|
||||||
Ordering continuous_ordering(graph.continuousKeySet());
|
Ordering continuous_ordering(graph.continuousKeySet());
|
||||||
|
|
@ -365,7 +365,7 @@ TEST(HybridEstimation, ProbabilityMultifrontal) {
|
||||||
// mode priors.
|
// mode priors.
|
||||||
Switching switching(K, between_sigma, measurement_sigma, measurements,
|
Switching switching(K, between_sigma, measurement_sigma, measurements,
|
||||||
"1/1 1/1");
|
"1/1 1/1");
|
||||||
auto graph = switching.linearizedFactorGraph;
|
auto graph = switching.linearizedFactorGraph();
|
||||||
|
|
||||||
// Get the tree of unnormalized probabilities for each mode sequence.
|
// Get the tree of unnormalized probabilities for each mode sequence.
|
||||||
AlgebraicDecisionTree<Key> expected_probPrimeTree = GetProbPrimeTree(graph);
|
AlgebraicDecisionTree<Key> expected_probPrimeTree = GetProbPrimeTree(graph);
|
||||||
|
|
|
||||||
|
|
@ -175,7 +175,7 @@ TEST(HybridBayesNet, Switching) {
|
||||||
Switching s(2, betweenSigma, priorSigma);
|
Switching s(2, betweenSigma, priorSigma);
|
||||||
|
|
||||||
// Check size of linearized factor graph
|
// Check size of linearized factor graph
|
||||||
const HybridGaussianFactorGraph &graph = s.linearizedFactorGraph;
|
const HybridGaussianFactorGraph &graph = s.linearizedFactorGraph();
|
||||||
EXPECT_LONGS_EQUAL(4, graph.size());
|
EXPECT_LONGS_EQUAL(4, graph.size());
|
||||||
|
|
||||||
// Create some continuous and discrete values
|
// Create some continuous and discrete values
|
||||||
|
|
@ -184,7 +184,7 @@ TEST(HybridBayesNet, Switching) {
|
||||||
const DiscreteValues modeZero{{M(0), 0}}, modeOne{{M(0), 1}};
|
const DiscreteValues modeZero{{M(0), 0}}, modeOne{{M(0), 1}};
|
||||||
|
|
||||||
// Get the hybrid gaussian factor and check it is as expected
|
// Get the hybrid gaussian factor and check it is as expected
|
||||||
auto hgf = std::dynamic_pointer_cast<HybridGaussianFactor>(graph.at(1));
|
auto hgf = std::dynamic_pointer_cast<HybridGaussianFactor>(graph.at(2));
|
||||||
CHECK(hgf);
|
CHECK(hgf);
|
||||||
|
|
||||||
// Get factors and scalars for both modes
|
// Get factors and scalars for both modes
|
||||||
|
|
@ -298,7 +298,7 @@ TEST(HybridBayesNet, Switching) {
|
||||||
factors_x1.push_back(
|
factors_x1.push_back(
|
||||||
factor); // Use the remaining factor from previous elimination
|
factor); // Use the remaining factor from previous elimination
|
||||||
factors_x1.push_back(
|
factors_x1.push_back(
|
||||||
graph.at(2)); // Add the factor for x1 from the original graph
|
graph.at(1)); // Add the factor for x1 from the original graph
|
||||||
|
|
||||||
// Test collectProductFactor for x1 clique
|
// Test collectProductFactor for x1 clique
|
||||||
auto productFactor_x1 = factors_x1.collectProductFactor();
|
auto productFactor_x1 = factors_x1.collectProductFactor();
|
||||||
|
|
@ -356,7 +356,7 @@ TEST(HybridGaussianFactorGraph, ErrorAndProbPrime) {
|
||||||
Switching s(3);
|
Switching s(3);
|
||||||
|
|
||||||
// Check size of linearized factor graph
|
// Check size of linearized factor graph
|
||||||
const HybridGaussianFactorGraph &graph = s.linearizedFactorGraph;
|
const HybridGaussianFactorGraph &graph = s.linearizedFactorGraph();
|
||||||
EXPECT_LONGS_EQUAL(7, graph.size());
|
EXPECT_LONGS_EQUAL(7, graph.size());
|
||||||
|
|
||||||
// Eliminate the graph
|
// Eliminate the graph
|
||||||
|
|
@ -383,16 +383,16 @@ TEST(HybridGaussianFactorGraph, ErrorAndProbPrime) {
|
||||||
TEST(HybridGaussianFactorGraph, DiscreteSelection) {
|
TEST(HybridGaussianFactorGraph, DiscreteSelection) {
|
||||||
Switching s(3);
|
Switching s(3);
|
||||||
|
|
||||||
HybridGaussianFactorGraph graph = s.linearizedFactorGraph;
|
HybridGaussianFactorGraph graph = s.linearizedFactorGraph();
|
||||||
|
|
||||||
DiscreteValues dv00{{M(0), 0}, {M(1), 0}};
|
DiscreteValues dv00{{M(0), 0}, {M(1), 0}};
|
||||||
GaussianFactorGraph continuous_00 = graph(dv00);
|
GaussianFactorGraph continuous_00 = graph(dv00);
|
||||||
GaussianFactorGraph expected_00;
|
GaussianFactorGraph expected_00;
|
||||||
expected_00.push_back(JacobianFactor(X(0), I_1x1 * 10, Vector1(-10)));
|
expected_00.push_back(JacobianFactor(X(0), I_1x1 * 10, Vector1(-10)));
|
||||||
expected_00.push_back(JacobianFactor(X(0), -I_1x1, X(1), I_1x1, Vector1(-1)));
|
|
||||||
expected_00.push_back(JacobianFactor(X(1), -I_1x1, X(2), I_1x1, Vector1(-1)));
|
|
||||||
expected_00.push_back(JacobianFactor(X(1), I_1x1 * 10, Vector1(-10)));
|
expected_00.push_back(JacobianFactor(X(1), I_1x1 * 10, Vector1(-10)));
|
||||||
expected_00.push_back(JacobianFactor(X(2), I_1x1 * 10, Vector1(-10)));
|
expected_00.push_back(JacobianFactor(X(2), I_1x1 * 10, Vector1(-10)));
|
||||||
|
expected_00.push_back(JacobianFactor(X(0), -I_1x1, X(1), I_1x1, Vector1(-1)));
|
||||||
|
expected_00.push_back(JacobianFactor(X(1), -I_1x1, X(2), I_1x1, Vector1(-1)));
|
||||||
|
|
||||||
EXPECT(assert_equal(expected_00, continuous_00));
|
EXPECT(assert_equal(expected_00, continuous_00));
|
||||||
|
|
||||||
|
|
@ -400,10 +400,10 @@ TEST(HybridGaussianFactorGraph, DiscreteSelection) {
|
||||||
GaussianFactorGraph continuous_01 = graph(dv01);
|
GaussianFactorGraph continuous_01 = graph(dv01);
|
||||||
GaussianFactorGraph expected_01;
|
GaussianFactorGraph expected_01;
|
||||||
expected_01.push_back(JacobianFactor(X(0), I_1x1 * 10, Vector1(-10)));
|
expected_01.push_back(JacobianFactor(X(0), I_1x1 * 10, Vector1(-10)));
|
||||||
expected_01.push_back(JacobianFactor(X(0), -I_1x1, X(1), I_1x1, Vector1(-1)));
|
|
||||||
expected_01.push_back(JacobianFactor(X(1), -I_1x1, X(2), I_1x1, Vector1(-0)));
|
|
||||||
expected_01.push_back(JacobianFactor(X(1), I_1x1 * 10, Vector1(-10)));
|
expected_01.push_back(JacobianFactor(X(1), I_1x1 * 10, Vector1(-10)));
|
||||||
expected_01.push_back(JacobianFactor(X(2), I_1x1 * 10, Vector1(-10)));
|
expected_01.push_back(JacobianFactor(X(2), I_1x1 * 10, Vector1(-10)));
|
||||||
|
expected_01.push_back(JacobianFactor(X(0), -I_1x1, X(1), I_1x1, Vector1(-1)));
|
||||||
|
expected_01.push_back(JacobianFactor(X(1), -I_1x1, X(2), I_1x1, Vector1(-0)));
|
||||||
|
|
||||||
EXPECT(assert_equal(expected_01, continuous_01));
|
EXPECT(assert_equal(expected_01, continuous_01));
|
||||||
|
|
||||||
|
|
@ -411,10 +411,10 @@ TEST(HybridGaussianFactorGraph, DiscreteSelection) {
|
||||||
GaussianFactorGraph continuous_10 = graph(dv10);
|
GaussianFactorGraph continuous_10 = graph(dv10);
|
||||||
GaussianFactorGraph expected_10;
|
GaussianFactorGraph expected_10;
|
||||||
expected_10.push_back(JacobianFactor(X(0), I_1x1 * 10, Vector1(-10)));
|
expected_10.push_back(JacobianFactor(X(0), I_1x1 * 10, Vector1(-10)));
|
||||||
expected_10.push_back(JacobianFactor(X(0), -I_1x1, X(1), I_1x1, Vector1(-0)));
|
|
||||||
expected_10.push_back(JacobianFactor(X(1), -I_1x1, X(2), I_1x1, Vector1(-1)));
|
|
||||||
expected_10.push_back(JacobianFactor(X(1), I_1x1 * 10, Vector1(-10)));
|
expected_10.push_back(JacobianFactor(X(1), I_1x1 * 10, Vector1(-10)));
|
||||||
expected_10.push_back(JacobianFactor(X(2), I_1x1 * 10, Vector1(-10)));
|
expected_10.push_back(JacobianFactor(X(2), I_1x1 * 10, Vector1(-10)));
|
||||||
|
expected_10.push_back(JacobianFactor(X(0), -I_1x1, X(1), I_1x1, Vector1(-0)));
|
||||||
|
expected_10.push_back(JacobianFactor(X(1), -I_1x1, X(2), I_1x1, Vector1(-1)));
|
||||||
|
|
||||||
EXPECT(assert_equal(expected_10, continuous_10));
|
EXPECT(assert_equal(expected_10, continuous_10));
|
||||||
|
|
||||||
|
|
@ -422,16 +422,16 @@ TEST(HybridGaussianFactorGraph, DiscreteSelection) {
|
||||||
GaussianFactorGraph continuous_11 = graph(dv11);
|
GaussianFactorGraph continuous_11 = graph(dv11);
|
||||||
GaussianFactorGraph expected_11;
|
GaussianFactorGraph expected_11;
|
||||||
expected_11.push_back(JacobianFactor(X(0), I_1x1 * 10, Vector1(-10)));
|
expected_11.push_back(JacobianFactor(X(0), I_1x1 * 10, Vector1(-10)));
|
||||||
expected_11.push_back(JacobianFactor(X(0), -I_1x1, X(1), I_1x1, Vector1(-0)));
|
|
||||||
expected_11.push_back(JacobianFactor(X(1), -I_1x1, X(2), I_1x1, Vector1(-0)));
|
|
||||||
expected_11.push_back(JacobianFactor(X(1), I_1x1 * 10, Vector1(-10)));
|
expected_11.push_back(JacobianFactor(X(1), I_1x1 * 10, Vector1(-10)));
|
||||||
expected_11.push_back(JacobianFactor(X(2), I_1x1 * 10, Vector1(-10)));
|
expected_11.push_back(JacobianFactor(X(2), I_1x1 * 10, Vector1(-10)));
|
||||||
|
expected_11.push_back(JacobianFactor(X(0), -I_1x1, X(1), I_1x1, Vector1(-0)));
|
||||||
|
expected_11.push_back(JacobianFactor(X(1), -I_1x1, X(2), I_1x1, Vector1(-0)));
|
||||||
|
|
||||||
EXPECT(assert_equal(expected_11, continuous_11));
|
EXPECT(assert_equal(expected_11, continuous_11));
|
||||||
}
|
}
|
||||||
|
|
||||||
/* ************************************************************************* */
|
/* ************************************************************************* */
|
||||||
TEST(HybridGaussianFactorGraph, optimize) {
|
TEST(HybridGaussianFactorGraph, Optimize) {
|
||||||
HybridGaussianFactorGraph hfg;
|
HybridGaussianFactorGraph hfg;
|
||||||
|
|
||||||
hfg.add(JacobianFactor(X(0), I_3x3, Z_3x1));
|
hfg.add(JacobianFactor(X(0), I_3x3, Z_3x1));
|
||||||
|
|
@ -451,16 +451,16 @@ TEST(HybridGaussianFactorGraph, Conditionals) {
|
||||||
Switching switching(4);
|
Switching switching(4);
|
||||||
|
|
||||||
HybridGaussianFactorGraph hfg;
|
HybridGaussianFactorGraph hfg;
|
||||||
hfg.push_back(switching.linearizedFactorGraph.at(0)); // P(X0)
|
hfg.push_back(switching.linearUnaryFactors.at(0)); // P(X0)
|
||||||
Ordering ordering;
|
Ordering ordering;
|
||||||
ordering.push_back(X(0));
|
ordering.push_back(X(0));
|
||||||
HybridBayesNet::shared_ptr bayes_net = hfg.eliminateSequential(ordering);
|
HybridBayesNet::shared_ptr bayes_net = hfg.eliminateSequential(ordering);
|
||||||
|
|
||||||
HybridGaussianFactorGraph hfg2;
|
HybridGaussianFactorGraph hfg2;
|
||||||
hfg2.push_back(*bayes_net); // P(X0)
|
hfg2.push_back(*bayes_net); // P(X0)
|
||||||
hfg2.push_back(switching.linearizedFactorGraph.at(1)); // P(X0, X1 | M0)
|
hfg2.push_back(switching.linearBinaryFactors.at(0)); // P(X0, X1 | M0)
|
||||||
hfg2.push_back(switching.linearizedFactorGraph.at(2)); // P(X1, X2 | M1)
|
hfg2.push_back(switching.linearBinaryFactors.at(1)); // P(X1, X2 | M1)
|
||||||
hfg2.push_back(switching.linearizedFactorGraph.at(5)); // P(M1)
|
hfg2.push_back(switching.linearUnaryFactors.at(2)); // P(X2)
|
||||||
ordering += X(1), X(2), M(0), M(1);
|
ordering += X(1), X(2), M(0), M(1);
|
||||||
|
|
||||||
// Created product of first two factors and check eliminate:
|
// Created product of first two factors and check eliminate:
|
||||||
|
|
@ -510,13 +510,13 @@ TEST(HybridGaussianFactorGraph, IncrementalErrorTree) {
|
||||||
Switching s(4);
|
Switching s(4);
|
||||||
|
|
||||||
HybridGaussianFactorGraph graph;
|
HybridGaussianFactorGraph graph;
|
||||||
graph.push_back(s.linearizedFactorGraph.at(0)); // f(X0)
|
graph.push_back(s.linearUnaryFactors.at(0)); // f(X0)
|
||||||
graph.push_back(s.linearizedFactorGraph.at(1)); // f(X0, X1, M0)
|
graph.push_back(s.linearBinaryFactors.at(0)); // f(X0, X1, M0)
|
||||||
graph.push_back(s.linearizedFactorGraph.at(2)); // f(X1, X2, M1)
|
graph.push_back(s.linearBinaryFactors.at(1)); // f(X1, X2, M1)
|
||||||
graph.push_back(s.linearizedFactorGraph.at(4)); // f(X1)
|
graph.push_back(s.linearUnaryFactors.at(1)); // f(X1)
|
||||||
graph.push_back(s.linearizedFactorGraph.at(5)); // f(X2)
|
graph.push_back(s.linearUnaryFactors.at(2)); // f(X2)
|
||||||
graph.push_back(s.linearizedFactorGraph.at(7)); // f(M0)
|
graph.push_back(s.modeChain.at(0)); // f(M0)
|
||||||
graph.push_back(s.linearizedFactorGraph.at(8)); // f(M0, M1)
|
graph.push_back(s.modeChain.at(1)); // f(M0, M1)
|
||||||
|
|
||||||
HybridBayesNet::shared_ptr hybridBayesNet = graph.eliminateSequential();
|
HybridBayesNet::shared_ptr hybridBayesNet = graph.eliminateSequential();
|
||||||
EXPECT_LONGS_EQUAL(5, hybridBayesNet->size());
|
EXPECT_LONGS_EQUAL(5, hybridBayesNet->size());
|
||||||
|
|
@ -531,8 +531,8 @@ TEST(HybridGaussianFactorGraph, IncrementalErrorTree) {
|
||||||
|
|
||||||
graph = HybridGaussianFactorGraph();
|
graph = HybridGaussianFactorGraph();
|
||||||
graph.push_back(*hybridBayesNet);
|
graph.push_back(*hybridBayesNet);
|
||||||
graph.push_back(s.linearizedFactorGraph.at(3)); // f(X2, X3, M2)
|
graph.push_back(s.linearBinaryFactors.at(2)); // f(X2, X3, M2)
|
||||||
graph.push_back(s.linearizedFactorGraph.at(6)); // f(X3)
|
graph.push_back(s.linearUnaryFactors.at(3)); // f(X3)
|
||||||
|
|
||||||
hybridBayesNet = graph.eliminateSequential();
|
hybridBayesNet = graph.eliminateSequential();
|
||||||
EXPECT_LONGS_EQUAL(7, hybridBayesNet->size());
|
EXPECT_LONGS_EQUAL(7, hybridBayesNet->size());
|
||||||
|
|
|
||||||
|
|
@ -42,15 +42,15 @@ using symbol_shorthand::Z;
|
||||||
|
|
||||||
/* ****************************************************************************/
|
/* ****************************************************************************/
|
||||||
namespace switching3 {
|
namespace switching3 {
|
||||||
// ϕ(x0) ϕ(x0,x1,m0) ϕ(x1,x2,m1) ϕ(x1;z1) ϕ(x2;z2) ϕ(m0) ϕ(m0,m1)
|
// ϕ(x0) ϕ(x1;z1) ϕ(x2;z2) ϕ(x0,x1,m0) ϕ(x1,x2,m1) ϕ(m0) ϕ(m0,m1)
|
||||||
const Switching switching(3);
|
const Switching switching(3);
|
||||||
const HybridGaussianFactorGraph &lfg = switching.linearizedFactorGraph;
|
const HybridGaussianFactorGraph &lfg = switching.linearizedFactorGraph();
|
||||||
|
|
||||||
// First update graph: ϕ(x0) ϕ(x0,x1,m0) ϕ(m0)
|
// First update graph: ϕ(x0) ϕ(x0,x1,m0) ϕ(m0)
|
||||||
const HybridGaussianFactorGraph graph1{lfg.at(0), lfg.at(1), lfg.at(5)};
|
const HybridGaussianFactorGraph graph1{lfg.at(0), lfg.at(3), lfg.at(5)};
|
||||||
|
|
||||||
// Second update graph: ϕ(x1,x2,m1) ϕ(x1;z1) ϕ(x2;z2) ϕ(m0,m1)
|
// Second update graph: ϕ(x1,x2,m1) ϕ(x1;z1) ϕ(x2;z2) ϕ(m0,m1)
|
||||||
const HybridGaussianFactorGraph graph2{lfg.at(2), lfg.at(3), lfg.at(4),
|
const HybridGaussianFactorGraph graph2{lfg.at(4), lfg.at(1), lfg.at(2),
|
||||||
lfg.at(6)};
|
lfg.at(6)};
|
||||||
} // namespace switching3
|
} // namespace switching3
|
||||||
|
|
||||||
|
|
@ -108,7 +108,7 @@ TEST(HybridGaussianElimination, IncrementalInference) {
|
||||||
|
|
||||||
// Now we calculate the expected factors using full elimination
|
// Now we calculate the expected factors using full elimination
|
||||||
const auto [expectedHybridBayesTree, expectedRemainingGraph] =
|
const auto [expectedHybridBayesTree, expectedRemainingGraph] =
|
||||||
switching.linearizedFactorGraph.eliminatePartialMultifrontal(ordering);
|
switching.linearizedFactorGraph().eliminatePartialMultifrontal(ordering);
|
||||||
|
|
||||||
// The densities on X(0) should be the same
|
// The densities on X(0) should be the same
|
||||||
auto x0_conditional = dynamic_pointer_cast<HybridGaussianConditional>(
|
auto x0_conditional = dynamic_pointer_cast<HybridGaussianConditional>(
|
||||||
|
|
@ -162,15 +162,14 @@ TEST(HybridGaussianElimination, Approx_inference) {
|
||||||
HybridGaussianFactorGraph graph1;
|
HybridGaussianFactorGraph graph1;
|
||||||
|
|
||||||
// Add the 3 hybrid factors, x0-x1, x1-x2, x2-x3
|
// Add the 3 hybrid factors, x0-x1, x1-x2, x2-x3
|
||||||
for (size_t i = 1; i < 4; i++) {
|
for (size_t i = 0; i < 3; i++) {
|
||||||
graph1.push_back(switching.linearizedFactorGraph.at(i));
|
graph1.push_back(switching.linearBinaryFactors.at(i));
|
||||||
}
|
}
|
||||||
|
|
||||||
// Add the Gaussian factors, 1 prior on X(0),
|
// Add the Gaussian factors, 1 prior on x0,
|
||||||
// 3 measurements on X(1), X(2), X(3)
|
// 3 measurements on x1, x2, x3
|
||||||
graph1.push_back(switching.linearizedFactorGraph.at(0));
|
for (size_t i = 0; i <= 3; i++) {
|
||||||
for (size_t i = 4; i <= 7; i++) {
|
graph1.push_back(switching.linearUnaryFactors.at(i));
|
||||||
graph1.push_back(switching.linearizedFactorGraph.at(i));
|
|
||||||
}
|
}
|
||||||
|
|
||||||
// Create ordering.
|
// Create ordering.
|
||||||
|
|
@ -181,7 +180,7 @@ TEST(HybridGaussianElimination, Approx_inference) {
|
||||||
|
|
||||||
// Now we calculate the actual factors using full elimination
|
// Now we calculate the actual factors using full elimination
|
||||||
const auto [unPrunedHybridBayesTree, unPrunedRemainingGraph] =
|
const auto [unPrunedHybridBayesTree, unPrunedRemainingGraph] =
|
||||||
switching.linearizedFactorGraph.eliminatePartialMultifrontal(ordering);
|
switching.linearizedFactorGraph().eliminatePartialMultifrontal(ordering);
|
||||||
|
|
||||||
size_t maxNrLeaves = 5;
|
size_t maxNrLeaves = 5;
|
||||||
incrementalHybrid.update(graph1);
|
incrementalHybrid.update(graph1);
|
||||||
|
|
@ -266,15 +265,14 @@ TEST_DISABLED(HybridGaussianElimination, IncrementalApproximate) {
|
||||||
|
|
||||||
/***** Run Round 1 *****/
|
/***** Run Round 1 *****/
|
||||||
// Add the 3 hybrid factors, x0-x1, x1-x2, x2-x3
|
// Add the 3 hybrid factors, x0-x1, x1-x2, x2-x3
|
||||||
for (size_t i = 1; i < 4; i++) {
|
for (size_t i = 0; i < 3; i++) {
|
||||||
graph.push_back(switching.linearizedFactorGraph.at(i));
|
graph.push_back(switching.linearBinaryFactors.at(i));
|
||||||
}
|
}
|
||||||
|
|
||||||
// Add the Gaussian factors, 1 prior on X(0),
|
// Add the Gaussian factors, 1 prior on x0,
|
||||||
graph.push_back(switching.linearizedFactorGraph.at(0));
|
// 3 measurements on x1, x2, x3
|
||||||
// 3 measurements on X(1), X(2), X(3)
|
for (size_t i = 0; i <= 3; i++) {
|
||||||
for (size_t i = 5; i <= 7; i++) {
|
graph.push_back(switching.linearUnaryFactors.at(i));
|
||||||
graph.push_back(switching.linearizedFactorGraph.at(i));
|
|
||||||
}
|
}
|
||||||
|
|
||||||
// Run update with pruning
|
// Run update with pruning
|
||||||
|
|
@ -295,11 +293,9 @@ TEST_DISABLED(HybridGaussianElimination, IncrementalApproximate) {
|
||||||
5, incrementalHybrid[X(3)]->conditional()->asHybrid()->nrComponents());
|
5, incrementalHybrid[X(3)]->conditional()->asHybrid()->nrComponents());
|
||||||
|
|
||||||
/***** Run Round 2 *****/
|
/***** Run Round 2 *****/
|
||||||
graph.resize(0); // clear all factors
|
graph = HybridGaussianFactorGraph();
|
||||||
// Add hybrid factor X(3)-X(4)
|
graph.push_back(switching.linearBinaryFactors.at(3)); // hybrid x3-x4
|
||||||
graph.push_back(switching.linearizedFactorGraph.at(4));
|
graph.push_back(switching.linearUnaryFactors.at(4)); // x4
|
||||||
// Add measurement factor X(4)
|
|
||||||
graph.push_back(switching.linearizedFactorGraph.at(8));
|
|
||||||
|
|
||||||
// Run update with pruning a second time.
|
// Run update with pruning a second time.
|
||||||
incrementalHybrid.update(graph);
|
incrementalHybrid.update(graph);
|
||||||
|
|
|
||||||
|
|
@ -189,8 +189,10 @@ TEST(HybridGaussianProductFactor, AddPruned) {
|
||||||
product += prunedFactorB;
|
product += prunedFactorB;
|
||||||
EXPECT_LONGS_EQUAL(6, product.nrLeaves());
|
EXPECT_LONGS_EQUAL(6, product.nrLeaves());
|
||||||
|
|
||||||
|
#ifdef GTSAM_DT_MERGING
|
||||||
auto pruned = product.removeEmpty();
|
auto pruned = product.removeEmpty();
|
||||||
EXPECT_LONGS_EQUAL(5, pruned.nrLeaves());
|
EXPECT_LONGS_EQUAL(5, pruned.nrLeaves());
|
||||||
|
#endif
|
||||||
}
|
}
|
||||||
|
|
||||||
/* ************************************************************************* */
|
/* ************************************************************************* */
|
||||||
|
|
|
||||||
|
|
@ -249,7 +249,7 @@ TEST(HybridNonlinearFactorGraph, Switching) {
|
||||||
Switching self(3);
|
Switching self(3);
|
||||||
|
|
||||||
EXPECT_LONGS_EQUAL(7, self.nonlinearFactorGraph().size());
|
EXPECT_LONGS_EQUAL(7, self.nonlinearFactorGraph().size());
|
||||||
EXPECT_LONGS_EQUAL(7, self.linearizedFactorGraph.size());
|
EXPECT_LONGS_EQUAL(7, self.linearizedFactorGraph().size());
|
||||||
}
|
}
|
||||||
|
|
||||||
/****************************************************************************
|
/****************************************************************************
|
||||||
|
|
@ -276,7 +276,7 @@ TEST(HybridNonlinearFactorGraph, EliminationTree) {
|
||||||
for (size_t k = 0; k < self.K; k++) ordering.push_back(X(k));
|
for (size_t k = 0; k < self.K; k++) ordering.push_back(X(k));
|
||||||
|
|
||||||
// Create elimination tree.
|
// Create elimination tree.
|
||||||
HybridEliminationTree etree(self.linearizedFactorGraph, ordering);
|
HybridEliminationTree etree(self.linearizedFactorGraph(), ordering);
|
||||||
EXPECT_LONGS_EQUAL(1, etree.roots().size())
|
EXPECT_LONGS_EQUAL(1, etree.roots().size())
|
||||||
}
|
}
|
||||||
|
|
||||||
|
|
@ -286,12 +286,12 @@ TEST(HybridNonlinearFactorGraph, EliminationTree) {
|
||||||
TEST(GaussianElimination, Eliminate_x0) {
|
TEST(GaussianElimination, Eliminate_x0) {
|
||||||
Switching self(3);
|
Switching self(3);
|
||||||
|
|
||||||
// Gather factors on x1, has a simple Gaussian and a hybrid factor.
|
// Gather factors on x0, has a simple Gaussian and a hybrid factor.
|
||||||
HybridGaussianFactorGraph factors;
|
HybridGaussianFactorGraph factors;
|
||||||
// Add gaussian prior
|
// Add gaussian prior
|
||||||
factors.push_back(self.linearizedFactorGraph[0]);
|
factors.push_back(self.linearUnaryFactors[0]);
|
||||||
// Add first hybrid factor
|
// Add first hybrid factor
|
||||||
factors.push_back(self.linearizedFactorGraph[1]);
|
factors.push_back(self.linearBinaryFactors[0]);
|
||||||
|
|
||||||
// Eliminate x0
|
// Eliminate x0
|
||||||
const Ordering ordering{X(0)};
|
const Ordering ordering{X(0)};
|
||||||
|
|
@ -313,8 +313,8 @@ TEST(HybridsGaussianElimination, Eliminate_x1) {
|
||||||
|
|
||||||
// Gather factors on x1, will be two hybrid factors (with x0 and x2, resp.).
|
// Gather factors on x1, will be two hybrid factors (with x0 and x2, resp.).
|
||||||
HybridGaussianFactorGraph factors;
|
HybridGaussianFactorGraph factors;
|
||||||
factors.push_back(self.linearizedFactorGraph[1]); // involves m0
|
factors.push_back(self.linearBinaryFactors[0]); // involves m0
|
||||||
factors.push_back(self.linearizedFactorGraph[2]); // involves m1
|
factors.push_back(self.linearBinaryFactors[1]); // involves m1
|
||||||
|
|
||||||
// Eliminate x1
|
// Eliminate x1
|
||||||
const Ordering ordering{X(1)};
|
const Ordering ordering{X(1)};
|
||||||
|
|
@ -349,7 +349,7 @@ GaussianFactorGraph::shared_ptr batchGFG(double between,
|
||||||
TEST(HybridGaussianElimination, EliminateHybrid_2_Variable) {
|
TEST(HybridGaussianElimination, EliminateHybrid_2_Variable) {
|
||||||
Switching self(2, 1.0, 0.1);
|
Switching self(2, 1.0, 0.1);
|
||||||
|
|
||||||
auto factors = self.linearizedFactorGraph;
|
auto factors = self.linearizedFactorGraph();
|
||||||
|
|
||||||
// Eliminate x0
|
// Eliminate x0
|
||||||
const Ordering ordering{X(0), X(1)};
|
const Ordering ordering{X(0), X(1)};
|
||||||
|
|
@ -380,15 +380,13 @@ TEST(HybridGaussianElimination, EliminateHybrid_2_Variable) {
|
||||||
TEST(HybridNonlinearFactorGraph, Partial_Elimination) {
|
TEST(HybridNonlinearFactorGraph, Partial_Elimination) {
|
||||||
Switching self(3);
|
Switching self(3);
|
||||||
|
|
||||||
auto linearizedFactorGraph = self.linearizedFactorGraph;
|
|
||||||
|
|
||||||
// Create ordering of only continuous variables.
|
// Create ordering of only continuous variables.
|
||||||
Ordering ordering;
|
Ordering ordering;
|
||||||
for (size_t k = 0; k < self.K; k++) ordering.push_back(X(k));
|
for (size_t k = 0; k < self.K; k++) ordering.push_back(X(k));
|
||||||
|
|
||||||
// Eliminate partially i.e. only continuous part.
|
// Eliminate partially i.e. only continuous part.
|
||||||
const auto [hybridBayesNet, remainingFactorGraph] =
|
const auto [hybridBayesNet, remainingFactorGraph] =
|
||||||
linearizedFactorGraph.eliminatePartialSequential(ordering);
|
self.linearizedFactorGraph().eliminatePartialSequential(ordering);
|
||||||
|
|
||||||
CHECK(hybridBayesNet);
|
CHECK(hybridBayesNet);
|
||||||
EXPECT_LONGS_EQUAL(3, hybridBayesNet->size());
|
EXPECT_LONGS_EQUAL(3, hybridBayesNet->size());
|
||||||
|
|
@ -444,11 +442,11 @@ TEST(HybridNonlinearFactorGraph, PrintErrors) {
|
||||||
// Get nonlinear factor graph and add linear factors to be holistic.
|
// Get nonlinear factor graph and add linear factors to be holistic.
|
||||||
// TODO(Frank): ???
|
// TODO(Frank): ???
|
||||||
HybridNonlinearFactorGraph fg = self.nonlinearFactorGraph();
|
HybridNonlinearFactorGraph fg = self.nonlinearFactorGraph();
|
||||||
fg.add(self.linearizedFactorGraph);
|
fg.add(self.linearizedFactorGraph());
|
||||||
|
|
||||||
// Optimize to get HybridValues
|
// Optimize to get HybridValues
|
||||||
HybridBayesNet::shared_ptr bn =
|
HybridBayesNet::shared_ptr bn =
|
||||||
self.linearizedFactorGraph.eliminateSequential();
|
self.linearizedFactorGraph().eliminateSequential();
|
||||||
HybridValues hv = bn->optimize();
|
HybridValues hv = bn->optimize();
|
||||||
|
|
||||||
// Print and verify
|
// Print and verify
|
||||||
|
|
@ -465,8 +463,6 @@ TEST(HybridNonlinearFactorGraph, PrintErrors) {
|
||||||
TEST(HybridNonlinearFactorGraph, Full_Elimination) {
|
TEST(HybridNonlinearFactorGraph, Full_Elimination) {
|
||||||
Switching self(3);
|
Switching self(3);
|
||||||
|
|
||||||
auto linearizedFactorGraph = self.linearizedFactorGraph;
|
|
||||||
|
|
||||||
// We first do a partial elimination
|
// We first do a partial elimination
|
||||||
DiscreteBayesNet discreteBayesNet;
|
DiscreteBayesNet discreteBayesNet;
|
||||||
|
|
||||||
|
|
@ -477,7 +473,7 @@ TEST(HybridNonlinearFactorGraph, Full_Elimination) {
|
||||||
|
|
||||||
// Eliminate partially.
|
// Eliminate partially.
|
||||||
const auto [hybridBayesNet_partial, remainingFactorGraph_partial] =
|
const auto [hybridBayesNet_partial, remainingFactorGraph_partial] =
|
||||||
linearizedFactorGraph.eliminatePartialSequential(ordering);
|
self.linearizedFactorGraph().eliminatePartialSequential(ordering);
|
||||||
|
|
||||||
DiscreteFactorGraph discrete_fg =
|
DiscreteFactorGraph discrete_fg =
|
||||||
remainingFactorGraph_partial->discreteFactors();
|
remainingFactorGraph_partial->discreteFactors();
|
||||||
|
|
@ -495,7 +491,7 @@ TEST(HybridNonlinearFactorGraph, Full_Elimination) {
|
||||||
|
|
||||||
// Eliminate partially.
|
// Eliminate partially.
|
||||||
HybridBayesNet::shared_ptr hybridBayesNet =
|
HybridBayesNet::shared_ptr hybridBayesNet =
|
||||||
linearizedFactorGraph.eliminateSequential(ordering);
|
self.linearizedFactorGraph().eliminateSequential(ordering);
|
||||||
|
|
||||||
CHECK(hybridBayesNet);
|
CHECK(hybridBayesNet);
|
||||||
EXPECT_LONGS_EQUAL(5, hybridBayesNet->size());
|
EXPECT_LONGS_EQUAL(5, hybridBayesNet->size());
|
||||||
|
|
@ -528,7 +524,7 @@ TEST(HybridNonlinearFactorGraph, Full_Elimination) {
|
||||||
TEST(HybridNonlinearFactorGraph, Printing) {
|
TEST(HybridNonlinearFactorGraph, Printing) {
|
||||||
Switching self(3);
|
Switching self(3);
|
||||||
|
|
||||||
auto linearizedFactorGraph = self.linearizedFactorGraph;
|
auto linearizedFactorGraph = self.linearizedFactorGraph();
|
||||||
|
|
||||||
// Create ordering.
|
// Create ordering.
|
||||||
Ordering ordering;
|
Ordering ordering;
|
||||||
|
|
@ -551,6 +547,24 @@ GaussianFactor:
|
||||||
No noise model
|
No noise model
|
||||||
|
|
||||||
Factor 1
|
Factor 1
|
||||||
|
GaussianFactor:
|
||||||
|
|
||||||
|
A[x1] = [
|
||||||
|
10
|
||||||
|
]
|
||||||
|
b = [ -10 ]
|
||||||
|
No noise model
|
||||||
|
|
||||||
|
Factor 2
|
||||||
|
GaussianFactor:
|
||||||
|
|
||||||
|
A[x2] = [
|
||||||
|
10
|
||||||
|
]
|
||||||
|
b = [ -10 ]
|
||||||
|
No noise model
|
||||||
|
|
||||||
|
Factor 3
|
||||||
HybridGaussianFactor:
|
HybridGaussianFactor:
|
||||||
Hybrid [x0 x1; m0]{
|
Hybrid [x0 x1; m0]{
|
||||||
Choice(m0)
|
Choice(m0)
|
||||||
|
|
@ -578,7 +592,7 @@ scalar: 0.918939
|
||||||
|
|
||||||
}
|
}
|
||||||
|
|
||||||
Factor 2
|
Factor 4
|
||||||
HybridGaussianFactor:
|
HybridGaussianFactor:
|
||||||
Hybrid [x1 x2; m1]{
|
Hybrid [x1 x2; m1]{
|
||||||
Choice(m1)
|
Choice(m1)
|
||||||
|
|
@ -606,24 +620,6 @@ scalar: 0.918939
|
||||||
|
|
||||||
}
|
}
|
||||||
|
|
||||||
Factor 3
|
|
||||||
GaussianFactor:
|
|
||||||
|
|
||||||
A[x1] = [
|
|
||||||
10
|
|
||||||
]
|
|
||||||
b = [ -10 ]
|
|
||||||
No noise model
|
|
||||||
|
|
||||||
Factor 4
|
|
||||||
GaussianFactor:
|
|
||||||
|
|
||||||
A[x2] = [
|
|
||||||
10
|
|
||||||
]
|
|
||||||
b = [ -10 ]
|
|
||||||
No noise model
|
|
||||||
|
|
||||||
Factor 5
|
Factor 5
|
||||||
DiscreteFactor:
|
DiscreteFactor:
|
||||||
P( m0 ):
|
P( m0 ):
|
||||||
|
|
@ -646,13 +642,35 @@ DiscreteFactor:
|
||||||
#else
|
#else
|
||||||
string expected_hybridFactorGraph = R"(
|
string expected_hybridFactorGraph = R"(
|
||||||
size: 7
|
size: 7
|
||||||
factor 0:
|
Factor 0
|
||||||
|
GaussianFactor:
|
||||||
|
|
||||||
A[x0] = [
|
A[x0] = [
|
||||||
10
|
10
|
||||||
]
|
]
|
||||||
b = [ -10 ]
|
b = [ -10 ]
|
||||||
No noise model
|
No noise model
|
||||||
factor 1:
|
|
||||||
|
Factor 1
|
||||||
|
GaussianFactor:
|
||||||
|
|
||||||
|
A[x1] = [
|
||||||
|
10
|
||||||
|
]
|
||||||
|
b = [ -10 ]
|
||||||
|
No noise model
|
||||||
|
|
||||||
|
Factor 2
|
||||||
|
GaussianFactor:
|
||||||
|
|
||||||
|
A[x2] = [
|
||||||
|
10
|
||||||
|
]
|
||||||
|
b = [ -10 ]
|
||||||
|
No noise model
|
||||||
|
|
||||||
|
Factor 3
|
||||||
|
HybridGaussianFactor:
|
||||||
Hybrid [x0 x1; m0]{
|
Hybrid [x0 x1; m0]{
|
||||||
Choice(m0)
|
Choice(m0)
|
||||||
0 Leaf :
|
0 Leaf :
|
||||||
|
|
@ -664,6 +682,7 @@ Hybrid [x0 x1; m0]{
|
||||||
]
|
]
|
||||||
b = [ -1 ]
|
b = [ -1 ]
|
||||||
No noise model
|
No noise model
|
||||||
|
scalar: 0.918939
|
||||||
|
|
||||||
1 Leaf :
|
1 Leaf :
|
||||||
A[x0] = [
|
A[x0] = [
|
||||||
|
|
@ -674,9 +693,12 @@ Hybrid [x0 x1; m0]{
|
||||||
]
|
]
|
||||||
b = [ -0 ]
|
b = [ -0 ]
|
||||||
No noise model
|
No noise model
|
||||||
|
scalar: 0.918939
|
||||||
|
|
||||||
}
|
}
|
||||||
factor 2:
|
|
||||||
|
Factor 4
|
||||||
|
HybridGaussianFactor:
|
||||||
Hybrid [x1 x2; m1]{
|
Hybrid [x1 x2; m1]{
|
||||||
Choice(m1)
|
Choice(m1)
|
||||||
0 Leaf :
|
0 Leaf :
|
||||||
|
|
@ -688,6 +710,7 @@ Hybrid [x1 x2; m1]{
|
||||||
]
|
]
|
||||||
b = [ -1 ]
|
b = [ -1 ]
|
||||||
No noise model
|
No noise model
|
||||||
|
scalar: 0.918939
|
||||||
|
|
||||||
1 Leaf :
|
1 Leaf :
|
||||||
A[x1] = [
|
A[x1] = [
|
||||||
|
|
@ -698,26 +721,21 @@ Hybrid [x1 x2; m1]{
|
||||||
]
|
]
|
||||||
b = [ -0 ]
|
b = [ -0 ]
|
||||||
No noise model
|
No noise model
|
||||||
|
scalar: 0.918939
|
||||||
|
|
||||||
}
|
}
|
||||||
factor 3:
|
|
||||||
A[x1] = [
|
Factor 5
|
||||||
10
|
DiscreteFactor:
|
||||||
]
|
P( m0 ):
|
||||||
b = [ -10 ]
|
|
||||||
No noise model
|
|
||||||
factor 4:
|
|
||||||
A[x2] = [
|
|
||||||
10
|
|
||||||
]
|
|
||||||
b = [ -10 ]
|
|
||||||
No noise model
|
|
||||||
factor 5: P( m0 ):
|
|
||||||
Choice(m0)
|
Choice(m0)
|
||||||
0 Leaf 0.5
|
0 Leaf 0.5
|
||||||
1 Leaf 0.5
|
1 Leaf 0.5
|
||||||
|
|
||||||
factor 6: P( m1 | m0 ):
|
|
||||||
|
Factor 6
|
||||||
|
DiscreteFactor:
|
||||||
|
P( m1 | m0 ):
|
||||||
Choice(m1)
|
Choice(m1)
|
||||||
0 Choice(m0)
|
0 Choice(m0)
|
||||||
0 0 Leaf 0.33333333
|
0 0 Leaf 0.33333333
|
||||||
|
|
@ -726,6 +744,7 @@ factor 6: P( m1 | m0 ):
|
||||||
1 0 Leaf 0.66666667
|
1 0 Leaf 0.66666667
|
||||||
1 1 Leaf 0.4
|
1 1 Leaf 0.4
|
||||||
|
|
||||||
|
|
||||||
)";
|
)";
|
||||||
#endif
|
#endif
|
||||||
|
|
||||||
|
|
|
||||||
|
|
@ -147,7 +147,7 @@ TEST(HybridNonlinearISAM, IncrementalInference) {
|
||||||
|
|
||||||
// Now we calculate the actual factors using full elimination
|
// Now we calculate the actual factors using full elimination
|
||||||
const auto [expectedHybridBayesTree, expectedRemainingGraph] =
|
const auto [expectedHybridBayesTree, expectedRemainingGraph] =
|
||||||
switching.linearizedFactorGraph
|
switching.linearizedFactorGraph()
|
||||||
.BaseEliminateable::eliminatePartialMultifrontal(ordering);
|
.BaseEliminateable::eliminatePartialMultifrontal(ordering);
|
||||||
|
|
||||||
// The densities on X(1) should be the same
|
// The densities on X(1) should be the same
|
||||||
|
|
@ -214,8 +214,6 @@ TEST(HybridNonlinearISAM, ApproxInference) {
|
||||||
initial.insert<double>(X(i), i + 1);
|
initial.insert<double>(X(i), i + 1);
|
||||||
}
|
}
|
||||||
|
|
||||||
// TODO(Frank): no mode chain?
|
|
||||||
|
|
||||||
// Create ordering.
|
// Create ordering.
|
||||||
Ordering ordering;
|
Ordering ordering;
|
||||||
for (size_t j = 0; j < 4; j++) {
|
for (size_t j = 0; j < 4; j++) {
|
||||||
|
|
@ -224,7 +222,7 @@ TEST(HybridNonlinearISAM, ApproxInference) {
|
||||||
|
|
||||||
// Now we calculate the actual factors using full elimination
|
// Now we calculate the actual factors using full elimination
|
||||||
const auto [unPrunedHybridBayesTree, unPrunedRemainingGraph] =
|
const auto [unPrunedHybridBayesTree, unPrunedRemainingGraph] =
|
||||||
switching.linearizedFactorGraph
|
switching.linearizedFactorGraph()
|
||||||
.BaseEliminateable::eliminatePartialMultifrontal(ordering);
|
.BaseEliminateable::eliminatePartialMultifrontal(ordering);
|
||||||
|
|
||||||
size_t maxNrLeaves = 5;
|
size_t maxNrLeaves = 5;
|
||||||
|
|
|
||||||
|
|
@ -130,7 +130,7 @@ TEST(HybridSerialization, HybridGaussianConditional) {
|
||||||
// Test HybridBayesNet serialization.
|
// Test HybridBayesNet serialization.
|
||||||
TEST(HybridSerialization, HybridBayesNet) {
|
TEST(HybridSerialization, HybridBayesNet) {
|
||||||
Switching s(2);
|
Switching s(2);
|
||||||
HybridBayesNet hbn = *(s.linearizedFactorGraph.eliminateSequential());
|
HybridBayesNet hbn = *(s.linearizedFactorGraph().eliminateSequential());
|
||||||
|
|
||||||
EXPECT(equalsObj<HybridBayesNet>(hbn));
|
EXPECT(equalsObj<HybridBayesNet>(hbn));
|
||||||
EXPECT(equalsXML<HybridBayesNet>(hbn));
|
EXPECT(equalsXML<HybridBayesNet>(hbn));
|
||||||
|
|
@ -141,7 +141,7 @@ TEST(HybridSerialization, HybridBayesNet) {
|
||||||
// Test HybridBayesTree serialization.
|
// Test HybridBayesTree serialization.
|
||||||
TEST(HybridSerialization, HybridBayesTree) {
|
TEST(HybridSerialization, HybridBayesTree) {
|
||||||
Switching s(2);
|
Switching s(2);
|
||||||
HybridBayesTree hbt = *(s.linearizedFactorGraph.eliminateMultifrontal());
|
HybridBayesTree hbt = *(s.linearizedFactorGraph().eliminateMultifrontal());
|
||||||
|
|
||||||
EXPECT(equalsObj<HybridBayesTree>(hbt));
|
EXPECT(equalsObj<HybridBayesTree>(hbt));
|
||||||
EXPECT(equalsXML<HybridBayesTree>(hbt));
|
EXPECT(equalsXML<HybridBayesTree>(hbt));
|
||||||
|
|
|
||||||
|
|
@ -26,16 +26,16 @@ namespace gtsam {
|
||||||
Vector AttitudeFactor::attitudeError(const Rot3& nRb,
|
Vector AttitudeFactor::attitudeError(const Rot3& nRb,
|
||||||
OptionalJacobian<2, 3> H) const {
|
OptionalJacobian<2, 3> H) const {
|
||||||
if (H) {
|
if (H) {
|
||||||
Matrix23 D_nRef_R;
|
Matrix23 D_nRotated_R;
|
||||||
Matrix22 D_e_nRef;
|
Matrix22 D_e_nRotated;
|
||||||
Unit3 nRef = nRb.rotate(bRef_, D_nRef_R);
|
Unit3 nRotated = nRb.rotate(bMeasured_, D_nRotated_R);
|
||||||
Vector e = nZ_.error(nRef, D_e_nRef);
|
Vector e = nRef_.error(nRotated, D_e_nRotated);
|
||||||
|
|
||||||
(*H) = D_e_nRef * D_nRef_R;
|
(*H) = D_e_nRotated * D_nRotated_R;
|
||||||
return e;
|
return e;
|
||||||
} else {
|
} else {
|
||||||
Unit3 nRef = nRb * bRef_;
|
Unit3 nRotated = nRb * bMeasured_;
|
||||||
return nZ_.error(nRef);
|
return nRef_.error(nRotated);
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
|
|
@ -44,8 +44,8 @@ void Rot3AttitudeFactor::print(const string& s,
|
||||||
const KeyFormatter& keyFormatter) const {
|
const KeyFormatter& keyFormatter) const {
|
||||||
cout << (s.empty() ? "" : s + " ") << "Rot3AttitudeFactor on "
|
cout << (s.empty() ? "" : s + " ") << "Rot3AttitudeFactor on "
|
||||||
<< keyFormatter(this->key()) << "\n";
|
<< keyFormatter(this->key()) << "\n";
|
||||||
nZ_.print(" measured direction in nav frame: ");
|
nRef_.print(" reference direction in nav frame: ");
|
||||||
bRef_.print(" reference direction in body frame: ");
|
bMeasured_.print(" measured direction in body frame: ");
|
||||||
this->noiseModel_->print(" noise model: ");
|
this->noiseModel_->print(" noise model: ");
|
||||||
}
|
}
|
||||||
|
|
||||||
|
|
@ -53,16 +53,16 @@ void Rot3AttitudeFactor::print(const string& s,
|
||||||
bool Rot3AttitudeFactor::equals(const NonlinearFactor& expected,
|
bool Rot3AttitudeFactor::equals(const NonlinearFactor& expected,
|
||||||
double tol) const {
|
double tol) const {
|
||||||
const This* e = dynamic_cast<const This*>(&expected);
|
const This* e = dynamic_cast<const This*>(&expected);
|
||||||
return e != nullptr && Base::equals(*e, tol) && this->nZ_.equals(e->nZ_, tol)
|
return e != nullptr && Base::equals(*e, tol) && this->nRef_.equals(e->nRef_, tol)
|
||||||
&& this->bRef_.equals(e->bRef_, tol);
|
&& this->bMeasured_.equals(e->bMeasured_, tol);
|
||||||
}
|
}
|
||||||
|
|
||||||
//***************************************************************************
|
//***************************************************************************
|
||||||
void Pose3AttitudeFactor::print(const string& s,
|
void Pose3AttitudeFactor::print(const string& s,
|
||||||
const KeyFormatter& keyFormatter) const {
|
const KeyFormatter& keyFormatter) const {
|
||||||
cout << s << "Pose3AttitudeFactor on " << keyFormatter(this->key()) << "\n";
|
cout << s << "Pose3AttitudeFactor on " << keyFormatter(this->key()) << "\n";
|
||||||
nZ_.print(" measured direction in nav frame: ");
|
nRef_.print(" reference direction in nav frame: ");
|
||||||
bRef_.print(" reference direction in body frame: ");
|
bMeasured_.print(" measured direction in body frame: ");
|
||||||
this->noiseModel_->print(" noise model: ");
|
this->noiseModel_->print(" noise model: ");
|
||||||
}
|
}
|
||||||
|
|
||||||
|
|
@ -70,8 +70,8 @@ void Pose3AttitudeFactor::print(const string& s,
|
||||||
bool Pose3AttitudeFactor::equals(const NonlinearFactor& expected,
|
bool Pose3AttitudeFactor::equals(const NonlinearFactor& expected,
|
||||||
double tol) const {
|
double tol) const {
|
||||||
const This* e = dynamic_cast<const This*>(&expected);
|
const This* e = dynamic_cast<const This*>(&expected);
|
||||||
return e != nullptr && Base::equals(*e, tol) && this->nZ_.equals(e->nZ_, tol)
|
return e != nullptr && Base::equals(*e, tol) && this->nRef_.equals(e->nRef_, tol)
|
||||||
&& this->bRef_.equals(e->bRef_, tol);
|
&& this->bMeasured_.equals(e->bMeasured_, tol);
|
||||||
}
|
}
|
||||||
|
|
||||||
//***************************************************************************
|
//***************************************************************************
|
||||||
|
|
|
||||||
|
|
@ -17,59 +17,69 @@
|
||||||
**/
|
**/
|
||||||
#pragma once
|
#pragma once
|
||||||
|
|
||||||
#include <gtsam/nonlinear/NonlinearFactor.h>
|
|
||||||
#include <gtsam/geometry/Pose3.h>
|
#include <gtsam/geometry/Pose3.h>
|
||||||
#include <gtsam/geometry/Unit3.h>
|
#include <gtsam/geometry/Unit3.h>
|
||||||
|
#include <gtsam/nonlinear/NonlinearFactor.h>
|
||||||
|
|
||||||
namespace gtsam {
|
namespace gtsam {
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* Base class for prior on attitude
|
* @class AttitudeFactor
|
||||||
* Example:
|
*
|
||||||
* - measurement is direction of gravity in body frame bF
|
* @brief Base class for an attitude factor that constrains the rotation between
|
||||||
* - reference is direction of gravity in navigation frame nG
|
* body and navigation frames.
|
||||||
* This factor will give zero error if nRb * bF == nG
|
*
|
||||||
|
* This factor enforces that when the measured direction in the body frame
|
||||||
|
* (e.g., from an IMU accelerometer) is rotated into the navigation frame using the
|
||||||
|
* rotation variable, it should align with a known reference direction in the
|
||||||
|
* navigation frame (e.g., gravity vector).
|
||||||
|
*
|
||||||
|
* Mathematically, the error is zero when:
|
||||||
|
* nRb * bMeasured == nRef
|
||||||
|
*
|
||||||
|
* This is useful for incorporating absolute orientation measurements into the
|
||||||
|
* factor graph.
|
||||||
|
*
|
||||||
* @ingroup navigation
|
* @ingroup navigation
|
||||||
*/
|
*/
|
||||||
class AttitudeFactor {
|
class AttitudeFactor {
|
||||||
|
|
||||||
protected:
|
protected:
|
||||||
|
Unit3 nRef_, bMeasured_; ///< Position measurement in
|
||||||
Unit3 nZ_, bRef_; ///< Position measurement in
|
|
||||||
|
|
||||||
public:
|
public:
|
||||||
|
|
||||||
/** default constructor - only use for serialization */
|
/** default constructor - only use for serialization */
|
||||||
AttitudeFactor() {
|
AttitudeFactor() {}
|
||||||
}
|
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* @brief Constructor
|
* @brief Constructor
|
||||||
* @param nZ measured direction in navigation frame
|
* @param nRef Reference direction in the navigation frame (e.g., gravity).
|
||||||
* @param bRef reference direction in body frame (default Z-axis in NED frame, i.e., [0; 0; 1])
|
* @param bMeasured Measured direction in the body frame (e.g., from IMU
|
||||||
|
* accelerometer). Default is Unit3(0, 0, 1).
|
||||||
*/
|
*/
|
||||||
AttitudeFactor(const Unit3& nZ, const Unit3& bRef = Unit3(0, 0, 1)) :
|
AttitudeFactor(const Unit3& nRef, const Unit3& bMeasured = Unit3(0, 0, 1))
|
||||||
nZ_(nZ), bRef_(bRef) {
|
: nRef_(nRef), bMeasured_(bMeasured) {}
|
||||||
}
|
|
||||||
|
|
||||||
/** vector of errors */
|
/** vector of errors */
|
||||||
Vector attitudeError(const Rot3& p,
|
Vector attitudeError(const Rot3& p, OptionalJacobian<2, 3> H = {}) const;
|
||||||
OptionalJacobian<2,3> H = {}) const;
|
|
||||||
|
|
||||||
const Unit3& nZ() const {
|
const Unit3& nRef() const { return nRef_; }
|
||||||
return nZ_;
|
const Unit3& bMeasured() const { return bMeasured_; }
|
||||||
}
|
|
||||||
const Unit3& bRef() const {
|
#ifdef GTSAM_ALLOW_DEPRECATED_SINCE_V43
|
||||||
return bRef_;
|
[[deprecated("Use nRef() instead")]]
|
||||||
}
|
const Unit3& nZ() const { return nRef_; }
|
||||||
|
|
||||||
|
[[deprecated("Use bMeasured() instead")]]
|
||||||
|
const Unit3& bRef() const { return bMeasured_; }
|
||||||
|
#endif
|
||||||
|
|
||||||
#ifdef GTSAM_ENABLE_BOOST_SERIALIZATION
|
#ifdef GTSAM_ENABLE_BOOST_SERIALIZATION
|
||||||
/** Serialization function */
|
/** Serialization function */
|
||||||
friend class boost::serialization::access;
|
friend class boost::serialization::access;
|
||||||
template <class ARCHIVE>
|
template <class ARCHIVE>
|
||||||
void serialize(ARCHIVE& ar, const unsigned int /*version*/) {
|
void serialize(ARCHIVE& ar, const unsigned int /*version*/) {
|
||||||
ar & boost::serialization::make_nvp("nZ_", nZ_);
|
ar& boost::serialization::make_nvp("nRef_", nRef_);
|
||||||
ar & boost::serialization::make_nvp("bRef_", bRef_);
|
ar& boost::serialization::make_nvp("bMeasured_", bMeasured_);
|
||||||
}
|
}
|
||||||
#endif
|
#endif
|
||||||
};
|
};
|
||||||
|
|
@ -78,12 +88,11 @@ public:
|
||||||
* Version of AttitudeFactor for Rot3
|
* Version of AttitudeFactor for Rot3
|
||||||
* @ingroup navigation
|
* @ingroup navigation
|
||||||
*/
|
*/
|
||||||
class GTSAM_EXPORT Rot3AttitudeFactor: public NoiseModelFactorN<Rot3>, public AttitudeFactor {
|
class GTSAM_EXPORT Rot3AttitudeFactor : public NoiseModelFactorN<Rot3>,
|
||||||
|
public AttitudeFactor {
|
||||||
typedef NoiseModelFactorN<Rot3> Base;
|
typedef NoiseModelFactorN<Rot3> Base;
|
||||||
|
|
||||||
public:
|
public:
|
||||||
|
|
||||||
// Provide access to the Matrix& version of evaluateError:
|
// Provide access to the Matrix& version of evaluateError:
|
||||||
using Base::evaluateError;
|
using Base::evaluateError;
|
||||||
|
|
||||||
|
|
@ -94,23 +103,20 @@ public:
|
||||||
typedef Rot3AttitudeFactor This;
|
typedef Rot3AttitudeFactor This;
|
||||||
|
|
||||||
/** default constructor - only use for serialization */
|
/** default constructor - only use for serialization */
|
||||||
Rot3AttitudeFactor() {
|
Rot3AttitudeFactor() {}
|
||||||
}
|
|
||||||
|
|
||||||
~Rot3AttitudeFactor() override {
|
~Rot3AttitudeFactor() override {}
|
||||||
}
|
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* @brief Constructor
|
* @brief Constructor
|
||||||
* @param key of the Rot3 variable that will be constrained
|
* @param key of the Rot3 variable that will be constrained
|
||||||
* @param nZ measured direction in navigation frame
|
* @param nRef reference direction in navigation frame
|
||||||
* @param model Gaussian noise model
|
* @param model Gaussian noise model
|
||||||
* @param bRef reference direction in body frame (default Z-axis)
|
* @param bMeasured measured direction in body frame (default Z-axis)
|
||||||
*/
|
*/
|
||||||
Rot3AttitudeFactor(Key key, const Unit3& nZ, const SharedNoiseModel& model,
|
Rot3AttitudeFactor(Key key, const Unit3& nRef, const SharedNoiseModel& model,
|
||||||
const Unit3& bRef = Unit3(0, 0, 1)) :
|
const Unit3& bMeasured = Unit3(0, 0, 1))
|
||||||
Base(model, key), AttitudeFactor(nZ, bRef) {
|
: Base(model, key), AttitudeFactor(nRef, bMeasured) {}
|
||||||
}
|
|
||||||
|
|
||||||
/// @return a deep copy of this factor
|
/// @return a deep copy of this factor
|
||||||
gtsam::NonlinearFactor::shared_ptr clone() const override {
|
gtsam::NonlinearFactor::shared_ptr clone() const override {
|
||||||
|
|
@ -123,7 +129,8 @@ public:
|
||||||
DefaultKeyFormatter) const override;
|
DefaultKeyFormatter) const override;
|
||||||
|
|
||||||
/** equals */
|
/** equals */
|
||||||
bool equals(const NonlinearFactor& expected, double tol = 1e-9) const override;
|
bool equals(const NonlinearFactor& expected,
|
||||||
|
double tol = 1e-9) const override;
|
||||||
|
|
||||||
/** vector of errors */
|
/** vector of errors */
|
||||||
Vector evaluateError(const Rot3& nRb, OptionalMatrixType H) const override {
|
Vector evaluateError(const Rot3& nRb, OptionalMatrixType H) const override {
|
||||||
|
|
@ -131,16 +138,16 @@ public:
|
||||||
}
|
}
|
||||||
|
|
||||||
private:
|
private:
|
||||||
|
|
||||||
#ifdef GTSAM_ENABLE_BOOST_SERIALIZATION
|
#ifdef GTSAM_ENABLE_BOOST_SERIALIZATION
|
||||||
/** Serialization function */
|
/** Serialization function */
|
||||||
friend class boost::serialization::access;
|
friend class boost::serialization::access;
|
||||||
template <class ARCHIVE>
|
template <class ARCHIVE>
|
||||||
void serialize(ARCHIVE& ar, const unsigned int /*version*/) {
|
void serialize(ARCHIVE& ar, const unsigned int /*version*/) {
|
||||||
// NoiseModelFactor1 instead of NoiseModelFactorN for backward compatibility
|
// NoiseModelFactor1 instead of NoiseModelFactorN for backward compatibility
|
||||||
ar & boost::serialization::make_nvp("NoiseModelFactor1",
|
ar& boost::serialization::make_nvp(
|
||||||
boost::serialization::base_object<Base>(*this));
|
"NoiseModelFactor1", boost::serialization::base_object<Base>(*this));
|
||||||
ar & boost::serialization::make_nvp("AttitudeFactor",
|
ar& boost::serialization::make_nvp(
|
||||||
|
"AttitudeFactor",
|
||||||
boost::serialization::base_object<AttitudeFactor>(*this));
|
boost::serialization::base_object<AttitudeFactor>(*this));
|
||||||
}
|
}
|
||||||
#endif
|
#endif
|
||||||
|
|
@ -150,7 +157,8 @@ public:
|
||||||
};
|
};
|
||||||
|
|
||||||
/// traits
|
/// traits
|
||||||
template<> struct traits<Rot3AttitudeFactor> : public Testable<Rot3AttitudeFactor> {};
|
template <>
|
||||||
|
struct traits<Rot3AttitudeFactor> : public Testable<Rot3AttitudeFactor> {};
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* Version of AttitudeFactor for Pose3
|
* Version of AttitudeFactor for Pose3
|
||||||
|
|
@ -158,11 +166,9 @@ template<> struct traits<Rot3AttitudeFactor> : public Testable<Rot3AttitudeFacto
|
||||||
*/
|
*/
|
||||||
class GTSAM_EXPORT Pose3AttitudeFactor : public NoiseModelFactorN<Pose3>,
|
class GTSAM_EXPORT Pose3AttitudeFactor : public NoiseModelFactorN<Pose3>,
|
||||||
public AttitudeFactor {
|
public AttitudeFactor {
|
||||||
|
|
||||||
typedef NoiseModelFactorN<Pose3> Base;
|
typedef NoiseModelFactorN<Pose3> Base;
|
||||||
|
|
||||||
public:
|
public:
|
||||||
|
|
||||||
// Provide access to the Matrix& version of evaluateError:
|
// Provide access to the Matrix& version of evaluateError:
|
||||||
using Base::evaluateError;
|
using Base::evaluateError;
|
||||||
|
|
||||||
|
|
@ -173,23 +179,20 @@ public:
|
||||||
typedef Pose3AttitudeFactor This;
|
typedef Pose3AttitudeFactor This;
|
||||||
|
|
||||||
/** default constructor - only use for serialization */
|
/** default constructor - only use for serialization */
|
||||||
Pose3AttitudeFactor() {
|
Pose3AttitudeFactor() {}
|
||||||
}
|
|
||||||
|
|
||||||
~Pose3AttitudeFactor() override {
|
~Pose3AttitudeFactor() override {}
|
||||||
}
|
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* @brief Constructor
|
* @brief Constructor
|
||||||
* @param key of the Pose3 variable that will be constrained
|
* @param key of the Pose3 variable that will be constrained
|
||||||
* @param nZ measured direction in navigation frame
|
* @param nRef reference direction in navigation frame
|
||||||
* @param model Gaussian noise model
|
* @param model Gaussian noise model
|
||||||
* @param bRef reference direction in body frame (default Z-axis)
|
* @param bMeasured measured direction in body frame (default Z-axis)
|
||||||
*/
|
*/
|
||||||
Pose3AttitudeFactor(Key key, const Unit3& nZ, const SharedNoiseModel& model,
|
Pose3AttitudeFactor(Key key, const Unit3& nRef, const SharedNoiseModel& model,
|
||||||
const Unit3& bRef = Unit3(0, 0, 1)) :
|
const Unit3& bMeasured = Unit3(0, 0, 1))
|
||||||
Base(model, key), AttitudeFactor(nZ, bRef) {
|
: Base(model, key), AttitudeFactor(nRef, bMeasured) {}
|
||||||
}
|
|
||||||
|
|
||||||
/// @return a deep copy of this factor
|
/// @return a deep copy of this factor
|
||||||
gtsam::NonlinearFactor::shared_ptr clone() const override {
|
gtsam::NonlinearFactor::shared_ptr clone() const override {
|
||||||
|
|
@ -202,7 +205,8 @@ public:
|
||||||
DefaultKeyFormatter) const override;
|
DefaultKeyFormatter) const override;
|
||||||
|
|
||||||
/** equals */
|
/** equals */
|
||||||
bool equals(const NonlinearFactor& expected, double tol = 1e-9) const override;
|
bool equals(const NonlinearFactor& expected,
|
||||||
|
double tol = 1e-9) const override;
|
||||||
|
|
||||||
/** vector of errors */
|
/** vector of errors */
|
||||||
Vector evaluateError(const Pose3& nTb, OptionalMatrixType H) const override {
|
Vector evaluateError(const Pose3& nTb, OptionalMatrixType H) const override {
|
||||||
|
|
@ -216,16 +220,16 @@ public:
|
||||||
}
|
}
|
||||||
|
|
||||||
private:
|
private:
|
||||||
|
|
||||||
#ifdef GTSAM_ENABLE_BOOST_SERIALIZATION
|
#ifdef GTSAM_ENABLE_BOOST_SERIALIZATION
|
||||||
/** Serialization function */
|
/** Serialization function */
|
||||||
friend class boost::serialization::access;
|
friend class boost::serialization::access;
|
||||||
template <class ARCHIVE>
|
template <class ARCHIVE>
|
||||||
void serialize(ARCHIVE& ar, const unsigned int /*version*/) {
|
void serialize(ARCHIVE& ar, const unsigned int /*version*/) {
|
||||||
// NoiseModelFactor1 instead of NoiseModelFactorN for backward compatibility
|
// NoiseModelFactor1 instead of NoiseModelFactorN for backward compatibility
|
||||||
ar & boost::serialization::make_nvp("NoiseModelFactor1",
|
ar& boost::serialization::make_nvp(
|
||||||
boost::serialization::base_object<Base>(*this));
|
"NoiseModelFactor1", boost::serialization::base_object<Base>(*this));
|
||||||
ar & boost::serialization::make_nvp("AttitudeFactor",
|
ar& boost::serialization::make_nvp(
|
||||||
|
"AttitudeFactor",
|
||||||
boost::serialization::base_object<AttitudeFactor>(*this));
|
boost::serialization::base_object<AttitudeFactor>(*this));
|
||||||
}
|
}
|
||||||
#endif
|
#endif
|
||||||
|
|
@ -235,7 +239,7 @@ public:
|
||||||
};
|
};
|
||||||
|
|
||||||
/// traits
|
/// traits
|
||||||
template<> struct traits<Pose3AttitudeFactor> : public Testable<Pose3AttitudeFactor> {};
|
template <>
|
||||||
|
struct traits<Pose3AttitudeFactor> : public Testable<Pose3AttitudeFactor> {};
|
||||||
} /// namespace gtsam
|
|
||||||
|
|
||||||
|
} // namespace gtsam
|
||||||
|
|
|
||||||
|
|
@ -0,0 +1,136 @@
|
||||||
|
# AttitudeFactor in GTSAM
|
||||||
|
|
||||||
|
[Cautionary note: this was generated from the source using ChatGPT but edited by Frank]
|
||||||
|
|
||||||
|
## Introduction
|
||||||
|
|
||||||
|
The `AttitudeFactor` in GTSAM is a factor that constrains the orientation (attitude) of a robot or sensor platform based on directional measurements. This is particularly useful in GPS-denied navigation contexts where orientation must be estimated from inertial sensors like accelerometers or magnetometers.
|
||||||
|
|
||||||
|
This document explains the mathematical foundation of the `AttitudeFactor` and guides users on how to use this factor effectively in GTSAM.
|
||||||
|
|
||||||
|
## Mathematical Foundation
|
||||||
|
|
||||||
|
### Concept
|
||||||
|
|
||||||
|
The `AttitudeFactor` constrains the rotation $\mathbf{R}_{nb}$ (from body frame $b$ to navigation frame $n$) such that a known reference direction in the navigation frame aligns with a measured direction in the body frame, when rotated. The factor enforces that:
|
||||||
|
|
||||||
|
$$
|
||||||
|
\text{nRef} \approx \mathbf{R}_{nb} \cdot \text{bMeasured}
|
||||||
|
$$
|
||||||
|
|
||||||
|
where:
|
||||||
|
|
||||||
|
- $\mathbf{R}_{nb}$ is the rotation matrix representing the orientation from body to navigation frame.
|
||||||
|
- $\text{bMeasured}$ is the measured direction in the body frame (e.g., the accelerometer reading).
|
||||||
|
- $\text{nRef}$ is the known reference direction in the navigation frame (e.g., the gravity vector in nav).
|
||||||
|
|
||||||
|
### Error Function
|
||||||
|
|
||||||
|
The error function computes the angular difference between the rotated reference direction and the measured direction:
|
||||||
|
|
||||||
|
$$
|
||||||
|
\mathbf{e} = \text{error}(\text{nRef}, \mathbf{R}_{nb} \cdot \text{bMeasured})
|
||||||
|
$$
|
||||||
|
|
||||||
|
This error is minimal (zero) when the rotated body reference direction aligns perfectly with the measured navigation direction.
|
||||||
|
|
||||||
|
The error is computed internally using the `attitudeError` function:
|
||||||
|
|
||||||
|
```cpp
|
||||||
|
Vector AttitudeFactor::attitudeError(const Rot3& nRb) const {
|
||||||
|
Unit3 nRotated = nRb * bMeasured_;
|
||||||
|
return nRef_.error(nRotated);
|
||||||
|
}
|
||||||
|
```
|
||||||
|
|
||||||
|
#### Explanation:
|
||||||
|
- The function computes the rotated measurement `nRotated` and then the error between `nRef` and `nRotated`.
|
||||||
|
- `nRef_.error(nRotated)` is a 2D vector-valued error between two directions, defined in [Unit3.h](../geometry/Unit3.h).
|
||||||
|
|
||||||
|
### Jacobians
|
||||||
|
|
||||||
|
For optimization, the $2 \times 3$ Jacobian of the error function with respect to the rotation parameters is required. The Jacobian is computed using chain rule differentiation, involving the derivative of the rotated vector with respect to the rotation parameters and the derivative of the error with respect to the rotated vector.
|
||||||
|
|
||||||
|
Note the Jacobian for this specific error function vanishes 180 degrees away from the true direction, and the factor is only expected to behave well when the `nRb` value is initialized in the correct hemisphere.
|
||||||
|
|
||||||
|
## Usage in GTSAM
|
||||||
|
|
||||||
|
### Including the Header
|
||||||
|
|
||||||
|
Include the `AttitudeFactor.h` header in your code:
|
||||||
|
|
||||||
|
```cpp
|
||||||
|
#include <gtsam/navigation/AttitudeFactor.h>
|
||||||
|
```
|
||||||
|
|
||||||
|
### Creating an Attitude Factor
|
||||||
|
|
||||||
|
You can create an attitude factor for either a `Rot3` (rotation only) or a `Pose3` (position and rotation) variable.
|
||||||
|
|
||||||
|
#### For `Rot3` Variables
|
||||||
|
|
||||||
|
```cpp
|
||||||
|
// Define keys
|
||||||
|
gtsam::Key rotKey = ...;
|
||||||
|
|
||||||
|
// Known reference direction in navigation frame (e.g., gravity)
|
||||||
|
gtsam::Unit3 nRef(0, 0, -1); // Assuming gravity points down in navigation frame
|
||||||
|
|
||||||
|
// Measured direction in body frame (e.g., accelerometer direction)
|
||||||
|
gtsam::Unit3 bMeasured(0, 0, 9.8); // will be normalized automatically
|
||||||
|
|
||||||
|
// Noise model
|
||||||
|
auto noiseModel = gtsam::noiseModel::Isotropic::Sigma(2, 0.1); // 2D error, sigma = 0.1
|
||||||
|
|
||||||
|
// Add to factor graph
|
||||||
|
gtsam::NonlinearFactorGraph graph;
|
||||||
|
graph.emplace_shared<Rot3AttitudeFactor>(rotKey, nRef, noiseModel, bMeasured);
|
||||||
|
```
|
||||||
|
|
||||||
|
#### For `Pose3` Variables
|
||||||
|
|
||||||
|
There is also a `Pose3AttitudeFactor` that automatically extracts the rotation from the pose, taking into account the chain rule for this operation so the Jacobians with respect to pose are correct.
|
||||||
|
The same caveat about vanishing Jacobian holds.
|
||||||
|
|
||||||
|
```cpp
|
||||||
|
// Define keys
|
||||||
|
gtsam::Key poseKey = ...;
|
||||||
|
|
||||||
|
// Known reference direction in navigation frame (e.g., gravity)
|
||||||
|
gtsam::Unit3 nRef(0, 0, -1); // Assuming gravity points down in navigation frame
|
||||||
|
|
||||||
|
// Measured direction in body frame (e.g., accelerometer direction)
|
||||||
|
gtsam::Unit3 bMeasured(0, 0, 9.8); // will be normalized automatically
|
||||||
|
|
||||||
|
// Noise model
|
||||||
|
auto noiseModel = gtsam::noiseModel::Isotropic::Sigma(2, 0.1);
|
||||||
|
|
||||||
|
// Add to factor graph
|
||||||
|
gtsam::NonlinearFactorGraph graph;
|
||||||
|
graph.emplace_shared<Pose3AttitudeFactor>(poseKey, nRef, noiseModel, bMeasured);
|
||||||
|
```
|
||||||
|
|
||||||
|
### Explanation of Parameters
|
||||||
|
|
||||||
|
- **Key**: The variable key in the factor graph corresponding to the `Rot3` or `Pose3` variable you are constraining.
|
||||||
|
- **nRef**: The known direction in the navigation frame. This is typically obtained from sensors like accelerometers or magnetometers.
|
||||||
|
- **bMeasured**: The measured direction in the body frame. By default, this is set to the Z-axis `[0; 0; 1]`, but it should match the direction your sensor measures. When constructing a `Unit3`, will be automatically normalized.
|
||||||
|
- **noiseModel**: The noise model representing the uncertainty in the measurement. Adjust the standard deviation based on the confidence in your measurements.
|
||||||
|
|
||||||
|
## Example in GPS-Denied Navigation
|
||||||
|
|
||||||
|
In GPS-denied environments, orientation estimation relies heavily on inertial measurements. By incorporating the `AttitudeFactor`, you can:
|
||||||
|
|
||||||
|
- Constrain the roll and pitch angles using gravity measurements from an accelerometer.
|
||||||
|
- Constrain the yaw angle using magnetic field measurements from a magnetometer (with caution due to magnetic disturbances).
|
||||||
|
|
||||||
|
This factor helps maintain an accurate orientation estimate over time, which is crucial for applications like drone flight, underwater vehicles, or indoor robotics.
|
||||||
|
|
||||||
|
## Conclusion
|
||||||
|
|
||||||
|
The `AttitudeFactor` is a useful tool in GTSAM for incorporating orientation measurements into your factor graph. By aligning a measured direction in the body frame with a known reference direction in the navigation frame, it provides a constraint that improves the estimation of the rotation variable. Correct understanding and usage of this factor enhance the performance of navigation algorithms, especially in challenging environments where GPS is unavailable.
|
||||||
|
|
||||||
|
# References
|
||||||
|
|
||||||
|
- [GTSAM Documentation](https://gtsam.org/)
|
||||||
|
- [Unit3 Class Reference](https://gtsam.org/doxygen/)
|
||||||
|
|
@ -37,13 +37,9 @@ typedef ManifoldPreintegration PreintegrationType;
|
||||||
|
|
||||||
/*
|
/*
|
||||||
* If you are using the factor, please cite:
|
* If you are using the factor, please cite:
|
||||||
* L. Carlone, Z. Kira, C. Beall, V. Indelman, F. Dellaert, "Eliminating
|
* Christian Forster, Luca Carlone, Frank Dellaert, and Davide Scaramuzza,
|
||||||
* conditionally independent sets in factor graphs: a unifying perspective based
|
* "On-Manifold Preintegration for Real-Time Visual-Inertial Odometry", IEEE
|
||||||
* on smart factors", Int. Conf. on Robotics and Automation (ICRA), 2014.
|
* Transactions on Robotics, 2017.
|
||||||
*
|
|
||||||
* C. Forster, L. Carlone, F. Dellaert, D. Scaramuzza, "IMU Preintegration on
|
|
||||||
* Manifold for Efficient Visual-Inertial Maximum-a-Posteriori Estimation",
|
|
||||||
* Robotics: Science and Systems (RSS), 2015.
|
|
||||||
*
|
*
|
||||||
* REFERENCES:
|
* REFERENCES:
|
||||||
* [1] G.S. Chirikjian, "Stochastic Models, Information Theory, and Lie Groups",
|
* [1] G.S. Chirikjian, "Stochastic Models, Information Theory, and Lie Groups",
|
||||||
|
|
@ -54,8 +50,8 @@ typedef ManifoldPreintegration PreintegrationType;
|
||||||
* [3] L. Carlone, S. Williams, R. Roberts, "Preintegrated IMU factor:
|
* [3] L. Carlone, S. Williams, R. Roberts, "Preintegrated IMU factor:
|
||||||
* Computation of the Jacobian Matrices", Tech. Report, 2013.
|
* Computation of the Jacobian Matrices", Tech. Report, 2013.
|
||||||
* Available in this repo as "PreintegratedIMUJacobians.pdf".
|
* Available in this repo as "PreintegratedIMUJacobians.pdf".
|
||||||
* [4] C. Forster, L. Carlone, F. Dellaert, D. Scaramuzza, "IMU Preintegration on
|
* [4] C. Forster, L. Carlone, F. Dellaert, D. Scaramuzza, "IMU Preintegration
|
||||||
* Manifold for Efficient Visual-Inertial Maximum-a-Posteriori Estimation",
|
* on Manifold for Efficient Visual-Inertial Maximum-a-Posteriori Estimation",
|
||||||
* Robotics: Science and Systems (RSS), 2015.
|
* Robotics: Science and Systems (RSS), 2015.
|
||||||
*/
|
*/
|
||||||
|
|
||||||
|
|
|
||||||
|
|
@ -0,0 +1,42 @@
|
||||||
|
# Navigation Factors
|
||||||
|
|
||||||
|
This directory contains factors related to navigation, including various IMU factors.
|
||||||
|
|
||||||
|
## IMU Factor:
|
||||||
|
|
||||||
|
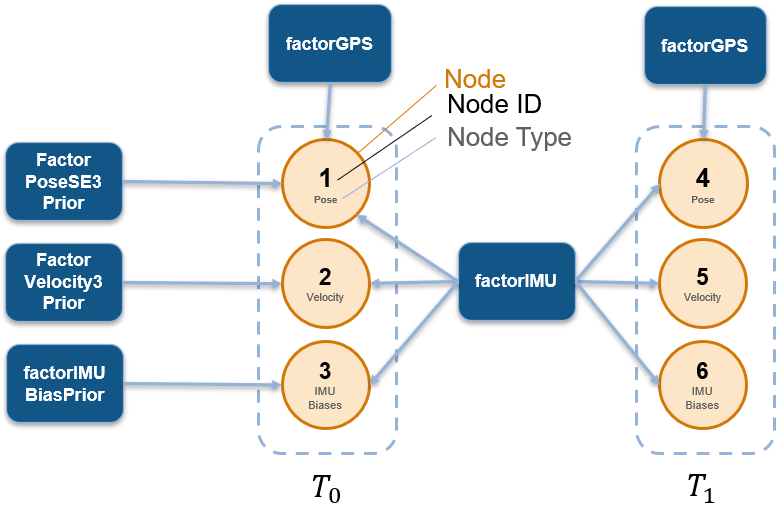
|
||||||
|
|
||||||
|
The `ImuFactor` is a 5-ways factor involving previous state (pose and velocity of
|
||||||
|
the vehicle at previous time step), current state (pose and velocity at
|
||||||
|
current time step), and the bias estimate.
|
||||||
|
Following the preintegration
|
||||||
|
scheme proposed in [2], the `ImuFactor` includes many IMU measurements, which
|
||||||
|
are "summarized" using the PreintegratedIMUMeasurements class.
|
||||||
|
The figure above, courtesy of [Mathworks' navigation toolbox](https://www.mathworks.com/help/nav/index.html), which are also using our work, shows the factor graph fragment for two time slices.
|
||||||
|
|
||||||
|
Note that this factor does not model "temporal consistency" of the biases
|
||||||
|
(which are usually slowly varying quantities), which is up to the caller.
|
||||||
|
See also `CombinedImuFactor` for a class that does this for you.
|
||||||
|
|
||||||
|
If you are using the factor, please cite:
|
||||||
|
> Christian Forster, Luca Carlone, Frank Dellaert, and Davide Scaramuzza, "On-Manifold Preintegration for Real-Time Visual-Inertial Odometry", IEEE Transactions on Robotics, 2017.
|
||||||
|
|
||||||
|
## REFERENCES:
|
||||||
|
1. G.S. Chirikjian, "Stochastic Models, Information Theory, and Lie Groups",
|
||||||
|
Volume 2, 2008.
|
||||||
|
2. T. Lupton and S.Sukkarieh, "Visual-Inertial-Aided Navigation for
|
||||||
|
High-Dynamic Motion in Built Environments Without Initial Conditions",
|
||||||
|
TRO, 28(1):61-76, 2012.
|
||||||
|
3. L. Carlone, S. Williams, R. Roberts, "Preintegrated IMU factor:
|
||||||
|
Computation of the Jacobian Matrices", Tech. Report, 2013.
|
||||||
|
Available in this repo as "PreintegratedIMUJacobians.pdf".
|
||||||
|
4. C. Forster, L. Carlone, F. Dellaert, D. Scaramuzza, "IMU Preintegration on
|
||||||
|
Manifold for Efficient Visual-Inertial Maximum-a-Posteriori Estimation",
|
||||||
|
Robotics: Science and Systems (RSS), 2015.
|
||||||
|
|
||||||
|
## The Attitude Factor
|
||||||
|
|
||||||
|
The `AttitudeFactor` in GTSAM is a factor that constrains the orientation (attitude) of a robot or sensor platform based on directional measurements. Both `Rot3` and `Pose3` versions are available.
|
||||||
|
|
||||||
|
Written up in detail with the help of ChatGPT [here](AttitudeFactor.md).
|
||||||
|
|
||||||
|
|
@ -256,34 +256,30 @@ virtual class AHRSFactor : gtsam::NonlinearFactor {
|
||||||
};
|
};
|
||||||
|
|
||||||
#include <gtsam/navigation/AttitudeFactor.h>
|
#include <gtsam/navigation/AttitudeFactor.h>
|
||||||
//virtual class AttitudeFactor : gtsam::NonlinearFactor {
|
virtual class Rot3AttitudeFactor : gtsam::NoiseModelFactor {
|
||||||
// AttitudeFactor(const Unit3& nZ, const Unit3& bRef);
|
Rot3AttitudeFactor(size_t key, const gtsam::Unit3& nRef, const gtsam::noiseModel::Diagonal* model,
|
||||||
// AttitudeFactor();
|
const gtsam::Unit3& bMeasured);
|
||||||
//};
|
Rot3AttitudeFactor(size_t key, const gtsam::Unit3& nRef, const gtsam::noiseModel::Diagonal* model);
|
||||||
virtual class Rot3AttitudeFactor : gtsam::NonlinearFactor{
|
|
||||||
Rot3AttitudeFactor(size_t key, const gtsam::Unit3& nZ, const gtsam::noiseModel::Diagonal* model,
|
|
||||||
const gtsam::Unit3& bRef);
|
|
||||||
Rot3AttitudeFactor(size_t key, const gtsam::Unit3& nZ, const gtsam::noiseModel::Diagonal* model);
|
|
||||||
Rot3AttitudeFactor();
|
Rot3AttitudeFactor();
|
||||||
void print(string s = "", const gtsam::KeyFormatter& keyFormatter =
|
void print(string s = "", const gtsam::KeyFormatter& keyFormatter =
|
||||||
gtsam::DefaultKeyFormatter) const;
|
gtsam::DefaultKeyFormatter) const;
|
||||||
bool equals(const gtsam::NonlinearFactor& expected, double tol) const;
|
bool equals(const gtsam::NonlinearFactor& expected, double tol) const;
|
||||||
gtsam::Unit3 nZ() const;
|
gtsam::Unit3 nRef() const;
|
||||||
gtsam::Unit3 bRef() const;
|
gtsam::Unit3 bMeasured() const;
|
||||||
};
|
};
|
||||||
|
|
||||||
virtual class Pose3AttitudeFactor : gtsam::NonlinearFactor {
|
virtual class Pose3AttitudeFactor : gtsam::NoiseModelFactor {
|
||||||
Pose3AttitudeFactor(size_t key, const gtsam::Unit3& nZ,
|
Pose3AttitudeFactor(size_t key, const gtsam::Unit3& nRef,
|
||||||
const gtsam::noiseModel::Diagonal* model,
|
const gtsam::noiseModel::Diagonal* model,
|
||||||
const gtsam::Unit3& bRef);
|
const gtsam::Unit3& bMeasured);
|
||||||
Pose3AttitudeFactor(size_t key, const gtsam::Unit3& nZ,
|
Pose3AttitudeFactor(size_t key, const gtsam::Unit3& nRef,
|
||||||
const gtsam::noiseModel::Diagonal* model);
|
const gtsam::noiseModel::Diagonal* model);
|
||||||
Pose3AttitudeFactor();
|
Pose3AttitudeFactor();
|
||||||
void print(string s = "", const gtsam::KeyFormatter& keyFormatter =
|
void print(string s = "", const gtsam::KeyFormatter& keyFormatter =
|
||||||
gtsam::DefaultKeyFormatter) const;
|
gtsam::DefaultKeyFormatter) const;
|
||||||
bool equals(const gtsam::NonlinearFactor& expected, double tol) const;
|
bool equals(const gtsam::NonlinearFactor& expected, double tol) const;
|
||||||
gtsam::Unit3 nZ() const;
|
gtsam::Unit3 nRef() const;
|
||||||
gtsam::Unit3 bRef() const;
|
gtsam::Unit3 bMeasured() const;
|
||||||
};
|
};
|
||||||
|
|
||||||
#include <gtsam/navigation/GPSFactor.h>
|
#include <gtsam/navigation/GPSFactor.h>
|
||||||
|
|
|
||||||
|
|
@ -11,18 +11,19 @@
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* @file testAttitudeFactor.cpp
|
* @file testAttitudeFactor.cpp
|
||||||
* @brief Unit test for Rot3AttitudeFactor
|
* @brief Unit test for AttitudeFactors (rot3 and Pose3 versions)
|
||||||
* @author Frank Dellaert
|
* @author Frank Dellaert
|
||||||
* @date January 22, 2014
|
* @date January 22, 2014
|
||||||
*/
|
*/
|
||||||
|
|
||||||
#include <gtsam/navigation/AttitudeFactor.h>
|
#include <CppUnitLite/TestHarness.h>
|
||||||
#include <gtsam/base/Testable.h>
|
#include <gtsam/base/Testable.h>
|
||||||
#include <gtsam/base/numericalDerivative.h>
|
#include <gtsam/base/numericalDerivative.h>
|
||||||
|
#include <gtsam/navigation/AttitudeFactor.h>
|
||||||
|
|
||||||
#include <functional>
|
#include <functional>
|
||||||
|
|
||||||
#include "gtsam/base/Matrix.h"
|
#include "gtsam/base/Matrix.h"
|
||||||
#include <CppUnitLite/TestHarness.h>
|
|
||||||
|
|
||||||
using namespace std::placeholders;
|
using namespace std::placeholders;
|
||||||
using namespace std;
|
using namespace std;
|
||||||
|
|
@ -30,18 +31,17 @@ using namespace gtsam;
|
||||||
|
|
||||||
// *************************************************************************
|
// *************************************************************************
|
||||||
TEST(Rot3AttitudeFactor, Constructor) {
|
TEST(Rot3AttitudeFactor, Constructor) {
|
||||||
|
|
||||||
// Example: pitch and roll of aircraft in an ENU Cartesian frame.
|
// Example: pitch and roll of aircraft in an ENU Cartesian frame.
|
||||||
// If pitch and roll are zero for an aerospace frame,
|
// If pitch and roll are zero for an aerospace frame,
|
||||||
// that means Z is pointing down, i.e., direction of Z = (0,0,-1)
|
// that means Z is pointing down, i.e., direction of Z = (0,0,-1)
|
||||||
Unit3 bZ(0, 0, 1); // reference direction is body Z axis
|
Unit3 bMeasured(0, 0, 1); // measured direction is body Z axis
|
||||||
Unit3 nDown(0, 0, -1); // down, in ENU navigation frame, is "measurement"
|
Unit3 nDown(0, 0, -1); // down, in ENU navigation frame, is "reference"
|
||||||
|
|
||||||
// Factor
|
// Factor
|
||||||
Key key(1);
|
Key key(1);
|
||||||
SharedNoiseModel model = noiseModel::Isotropic::Sigma(2, 0.25);
|
SharedNoiseModel model = noiseModel::Isotropic::Sigma(2, 0.25);
|
||||||
Rot3AttitudeFactor factor0(key, nDown, model);
|
Rot3AttitudeFactor factor0(key, nDown, model);
|
||||||
Rot3AttitudeFactor factor(key, nDown, model, bZ);
|
Rot3AttitudeFactor factor(key, nDown, model, bMeasured);
|
||||||
EXPECT(assert_equal(factor0, factor, 1e-5));
|
EXPECT(assert_equal(factor0, factor, 1e-5));
|
||||||
|
|
||||||
// Create a linearization point at the zero-error point
|
// Create a linearization point at the zero-error point
|
||||||
|
|
@ -81,18 +81,17 @@ TEST(Rot3AttitudeFactor, CopyAndMove) {
|
||||||
|
|
||||||
// *************************************************************************
|
// *************************************************************************
|
||||||
TEST(Pose3AttitudeFactor, Constructor) {
|
TEST(Pose3AttitudeFactor, Constructor) {
|
||||||
|
|
||||||
// Example: pitch and roll of aircraft in an ENU Cartesian frame.
|
// Example: pitch and roll of aircraft in an ENU Cartesian frame.
|
||||||
// If pitch and roll are zero for an aerospace frame,
|
// If pitch and roll are zero for an aerospace frame,
|
||||||
// that means Z is pointing down, i.e., direction of Z = (0,0,-1)
|
// that means Z is pointing down, i.e., direction of Z = (0,0,-1)
|
||||||
Unit3 bZ(0, 0, 1); // reference direction is body Z axis
|
Unit3 bMeasured(0, 0, 1); // measured direction is body Z axis
|
||||||
Unit3 nDown(0, 0, -1); // down, in ENU navigation frame, is "measurement"
|
Unit3 nDown(0, 0, -1); // down, in ENU navigation frame, is "reference"
|
||||||
|
|
||||||
// Factor
|
// Factor
|
||||||
Key key(1);
|
Key key(1);
|
||||||
SharedNoiseModel model = noiseModel::Isotropic::Sigma(2, 0.25);
|
SharedNoiseModel model = noiseModel::Isotropic::Sigma(2, 0.25);
|
||||||
Pose3AttitudeFactor factor0(key, nDown, model);
|
Pose3AttitudeFactor factor0(key, nDown, model);
|
||||||
Pose3AttitudeFactor factor(key, nDown, model, bZ);
|
Pose3AttitudeFactor factor(key, nDown, model, bMeasured);
|
||||||
EXPECT(assert_equal(factor0, factor, 1e-5));
|
EXPECT(assert_equal(factor0, factor, 1e-5));
|
||||||
|
|
||||||
// Create a linearization point at the zero-error point
|
// Create a linearization point at the zero-error point
|
||||||
|
|
|
||||||
|
|
@ -21,6 +21,8 @@
|
||||||
#include <gtsam/base/Manifold.h>
|
#include <gtsam/base/Manifold.h>
|
||||||
#include <gtsam/nonlinear/NonlinearOptimizer.h>
|
#include <gtsam/nonlinear/NonlinearOptimizer.h>
|
||||||
|
|
||||||
|
#include <stdexcept>
|
||||||
|
|
||||||
namespace gtsam {
|
namespace gtsam {
|
||||||
|
|
||||||
/// Fletcher-Reeves formula for computing β, the direction of steepest descent.
|
/// Fletcher-Reeves formula for computing β, the direction of steepest descent.
|
||||||
|
|
@ -247,6 +249,9 @@ std::tuple<V, int> nonlinearConjugateGradient(
|
||||||
case DirectionMethod::DaiYuan:
|
case DirectionMethod::DaiYuan:
|
||||||
beta = DaiYuan(currentGradient, prevGradient, direction);
|
beta = DaiYuan(currentGradient, prevGradient, direction);
|
||||||
break;
|
break;
|
||||||
|
default:
|
||||||
|
throw std::runtime_error(
|
||||||
|
"NonlinearConjugateGradientOptimizer: Invalid directionMethod");
|
||||||
}
|
}
|
||||||
|
|
||||||
direction = currentGradient + (beta * direction);
|
direction = currentGradient + (beta * direction);
|
||||||
|
|
|
||||||
Loading…
Reference in New Issue